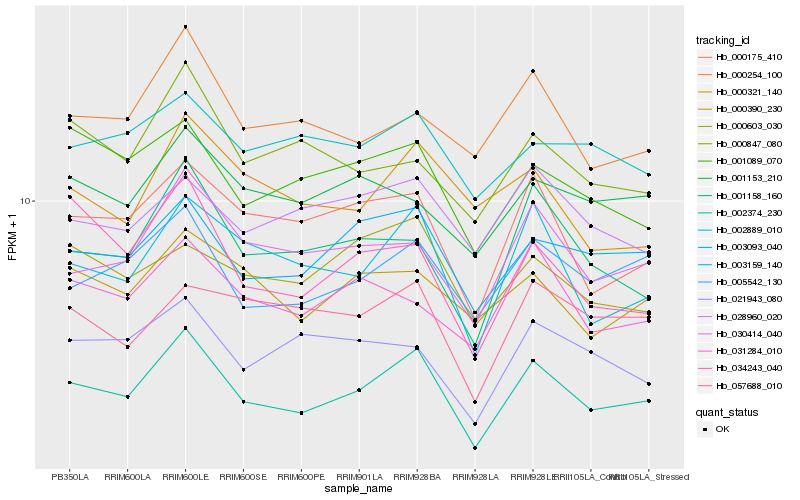
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_230 |
0.0 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 2 |
Hb_003093_040 |
0.0750769356 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_003159_140 |
0.0766409747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005542_130 |
0.0786239488 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 5 |
Hb_000175_410 |
0.0817379648 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_030414_040 |
0.0821728396 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001158_160 |
0.0843144335 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000254_100 |
0.0936658731 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 9 |
Hb_001089_070 |
0.0942406702 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 10 |
Hb_031284_010 |
0.0953321531 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 11 |
Hb_000390_230 |
0.0959160876 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 12 |
Hb_034243_040 |
0.096726558 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 13 |
Hb_057688_010 |
0.0981396104 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000603_030 |
0.0998155712 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 15 |
Hb_021943_080 |
0.1005873152 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_028960_020 |
0.1007362261 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000321_140 |
0.1016083391 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 18 |
Hb_001153_210 |
0.1017739887 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002889_010 |
0.1023253007 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 20 |
Hb_000847_080 |
0.1023995294 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |