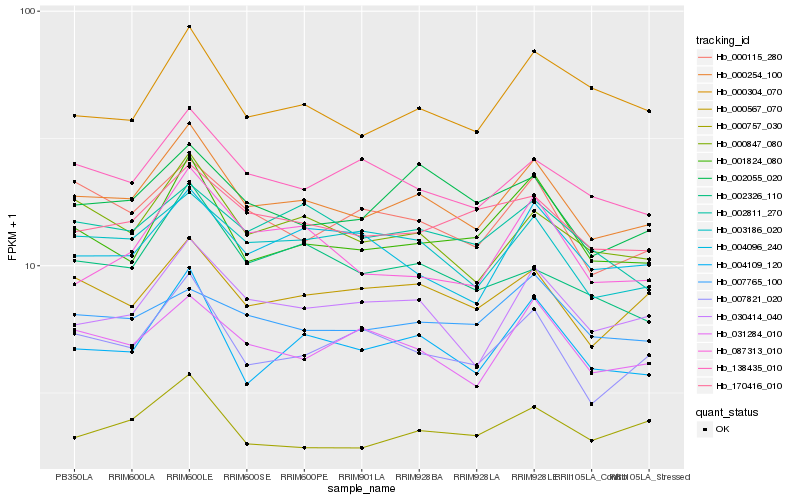
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000254_100 |
0.0 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 2 |
Hb_000847_080 |
0.058963797 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 3 |
Hb_004109_120 |
0.0610194662 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_030414_040 |
0.0621667871 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000567_070 |
0.0629593893 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 6 |
Hb_003186_020 |
0.0632690908 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001824_080 |
0.0649248395 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 8 |
Hb_007821_020 |
0.0671765655 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 9 |
Hb_138435_010 |
0.0680469416 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 10 |
Hb_170416_010 |
0.0691204207 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 11 |
Hb_031284_010 |
0.0703742382 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 12 |
Hb_002055_020 |
0.0716316015 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 13 |
Hb_000115_280 |
0.0721031307 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 14 |
Hb_000304_070 |
0.0738725625 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 15 |
Hb_002811_270 |
0.0757629103 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 16 |
Hb_000757_030 |
0.076750358 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 17 |
Hb_004096_240 |
0.076897351 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 18 |
Hb_002326_110 |
0.0773758687 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 19 |
Hb_007765_100 |
0.0779120424 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 20 |
Hb_087313_010 |
0.0787044668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |