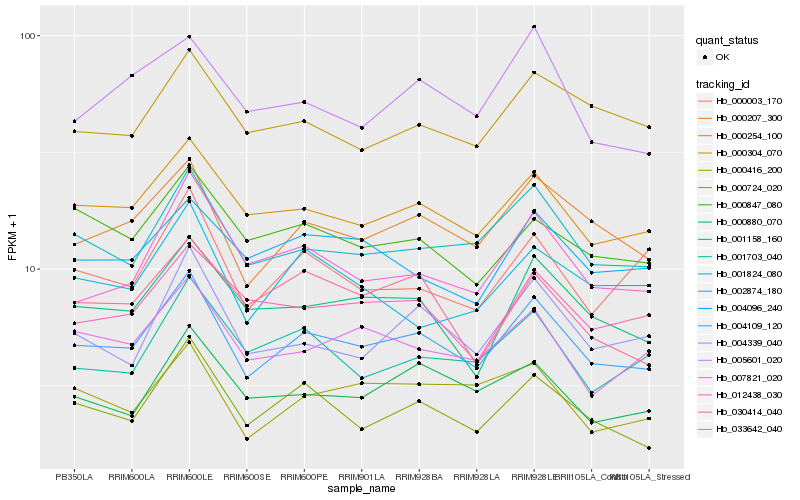
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_120 |
0.0 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000207_300 |
0.056598214 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000254_100 |
0.0610194662 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 4 |
Hb_001824_080 |
0.0629988904 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 5 |
Hb_000724_020 |
0.0763082727 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 6 |
Hb_001158_160 |
0.076685872 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004339_040 |
0.0797334128 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 8 |
Hb_012438_030 |
0.0810661957 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 9 |
Hb_007821_020 |
0.0824852415 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000304_070 |
0.0834050686 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 11 |
Hb_000880_070 |
0.0836732776 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 12 |
Hb_030414_040 |
0.0855047626 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004096_240 |
0.0873959336 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 14 |
Hb_000847_080 |
0.0879807622 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_005601_020 |
0.0884484641 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_000416_200 |
0.0885317863 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 17 |
Hb_001703_040 |
0.0901958791 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000003_170 |
0.0903230272 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 19 |
Hb_002874_180 |
0.0937164925 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_033642_040 |
0.0951249703 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |