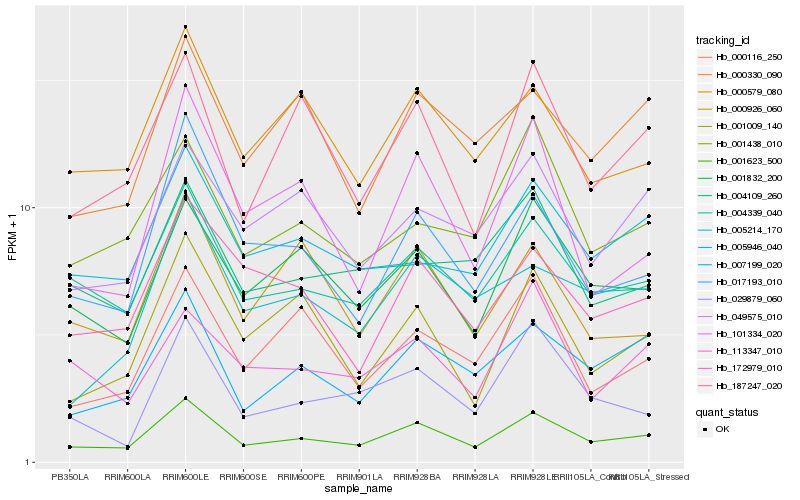
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_500 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 2 |
Hb_004339_040 |
0.0861044806 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 3 |
Hb_001832_200 |
0.0949756938 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007199_020 |
0.0981778159 |
- |
- |
hypothetical protein CICLE_v10028570mg [Citrus clementina] |
| 5 |
Hb_187247_020 |
0.1020069119 |
- |
- |
PREDICTED: uncharacterized protein LOC101220469 [Cucumis sativus] |
| 6 |
Hb_017193_010 |
0.1056948619 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000579_080 |
0.1089923553 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 8 |
Hb_005946_040 |
0.1167343931 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 9 |
Hb_172979_010 |
0.1172695479 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 10 |
Hb_001438_010 |
0.1175642452 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 11 |
Hb_101334_020 |
0.1192443442 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 12 |
Hb_001009_140 |
0.1195950803 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 13 |
Hb_000330_090 |
0.1198613954 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 14 |
Hb_049575_010 |
0.1203264424 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 15 |
Hb_000116_250 |
0.1216303254 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 16 |
Hb_000926_060 |
0.1236420453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_029879_060 |
0.1238493362 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 18 |
Hb_004109_260 |
0.125338962 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 19 |
Hb_005214_170 |
0.1262748457 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 20 |
Hb_113347_010 |
0.126633561 |
- |
- |
hypothetical protein CISIN_1g009336mg [Citrus sinensis] |