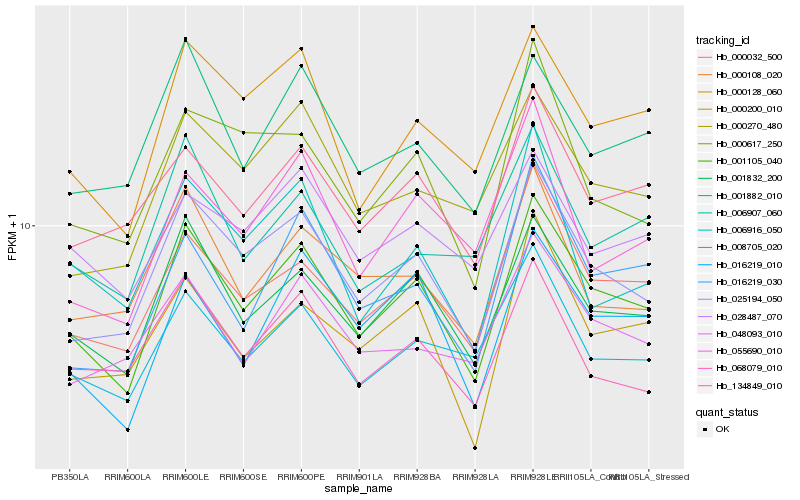
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001105_040 |
0.0 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001832_200 |
0.0615372227 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_025194_050 |
0.0617406156 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000032_500 |
0.0751811043 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 5 |
Hb_000128_060 |
0.0842760929 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 6 |
Hb_016219_010 |
0.0911159776 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 7 |
Hb_008705_020 |
0.0919470096 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_134849_010 |
0.0947589517 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 9 |
Hb_001882_010 |
0.0962974357 |
- |
- |
- |
| 10 |
Hb_016219_030 |
0.0970463017 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_000270_480 |
0.0983788381 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 12 |
Hb_000200_010 |
0.0998893239 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 13 |
Hb_000108_020 |
0.0999851832 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 14 |
Hb_068079_010 |
0.101122229 |
- |
- |
- |
| 15 |
Hb_000617_250 |
0.1028031869 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_055690_010 |
0.1048334765 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006907_060 |
0.1049712213 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_048093_010 |
0.1052444979 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_028487_070 |
0.1068255577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006916_050 |
0.10690085 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |