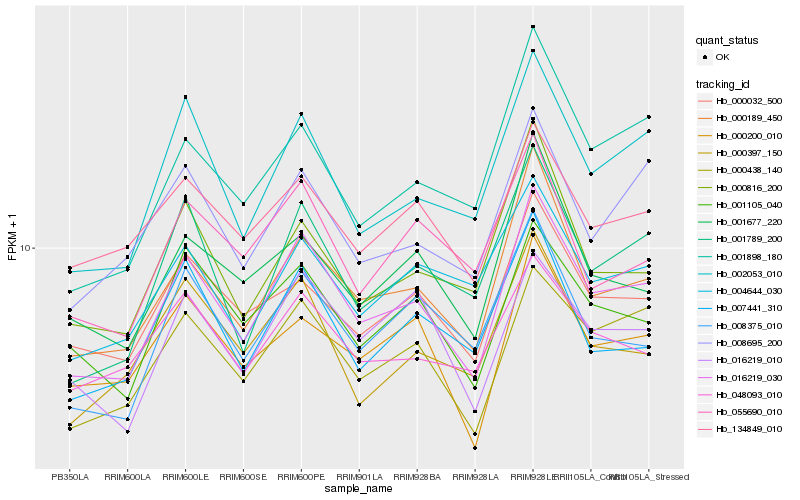
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016219_030 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000189_450 |
0.0748377267 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000438_140 |
0.0800174449 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 4 |
Hb_002053_010 |
0.0840197969 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_001789_200 |
0.0901304969 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000032_500 |
0.0956975794 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 7 |
Hb_001105_040 |
0.0970463017 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_016219_010 |
0.0996428787 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 9 |
Hb_001898_180 |
0.1014170224 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_008375_010 |
0.1017650764 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 11 |
Hb_008695_200 |
0.1018155447 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 12 |
Hb_000200_010 |
0.1029972725 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 13 |
Hb_007441_310 |
0.1043471067 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_048093_010 |
0.1046039743 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_001677_220 |
0.1053340742 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 16 |
Hb_000816_200 |
0.1055319379 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 17 |
Hb_004644_030 |
0.1066473152 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 18 |
Hb_134849_010 |
0.1077461121 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 19 |
Hb_000397_150 |
0.1081520567 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 20 |
Hb_055690_010 |
0.1105531905 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |