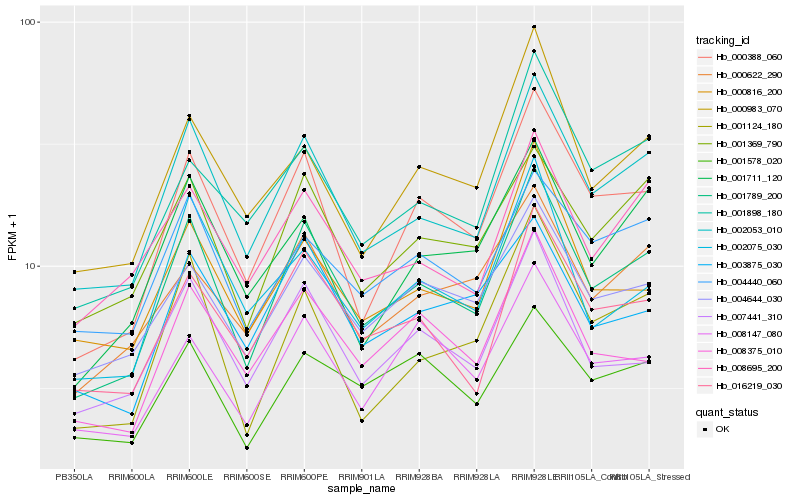
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_200 |
0.0 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002053_010 |
0.0525313082 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000388_060 |
0.0737747194 |
- |
- |
fructokinase [Manihot esculenta] |
| 4 |
Hb_001369_790 |
0.088831277 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 5 |
Hb_016219_030 |
0.0901304969 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_004644_030 |
0.0932078577 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 7 |
Hb_007441_310 |
0.0943699282 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001124_180 |
0.0961854066 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 9 |
Hb_004440_060 |
0.0995704745 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 10 |
Hb_008375_010 |
0.1002685071 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 11 |
Hb_000983_070 |
0.1014825417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001711_120 |
0.1017513891 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_008695_200 |
0.1019172542 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 14 |
Hb_008147_080 |
0.1022818034 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000816_200 |
0.103021035 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 16 |
Hb_000622_290 |
0.1034821394 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 17 |
Hb_001898_180 |
0.1038495899 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003875_030 |
0.1045237328 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 19 |
Hb_001578_020 |
0.1050084999 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 20 |
Hb_002075_030 |
0.105664804 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |