| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
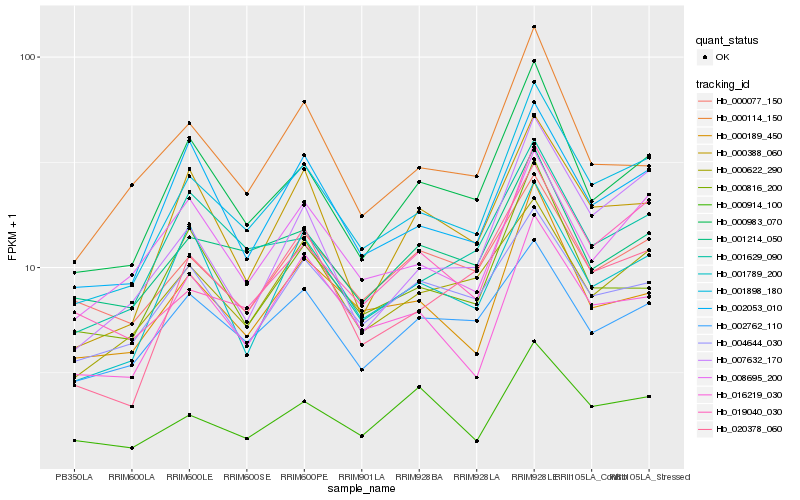
Hb_001898_180 |
0.0 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000983_070 |
0.0797493212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002053_010 |
0.0924038739 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_020378_060 |
0.0932837778 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 5 |
Hb_000189_450 |
0.0965473728 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_007632_170 |
0.0975538844 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 7 |
Hb_000622_290 |
0.0981138537 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 8 |
Hb_016219_030 |
0.1014170224 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000388_060 |
0.1027548928 |
- |
- |
fructokinase [Manihot esculenta] |
| 10 |
Hb_001629_090 |
0.1030220576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001214_050 |
0.1036336862 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001789_200 |
0.1038495899 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 13 |
Hb_008695_200 |
0.1039784625 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 14 |
Hb_019040_030 |
0.1048510795 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002762_110 |
0.105346835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004644_030 |
0.1073989964 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 17 |
Hb_000114_150 |
0.1111679913 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_000816_200 |
0.1124195008 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 19 |
Hb_000077_150 |
0.1128971108 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 20 |
Hb_000914_100 |
0.1155638779 |
- |
- |
unknown [Glycine max] |