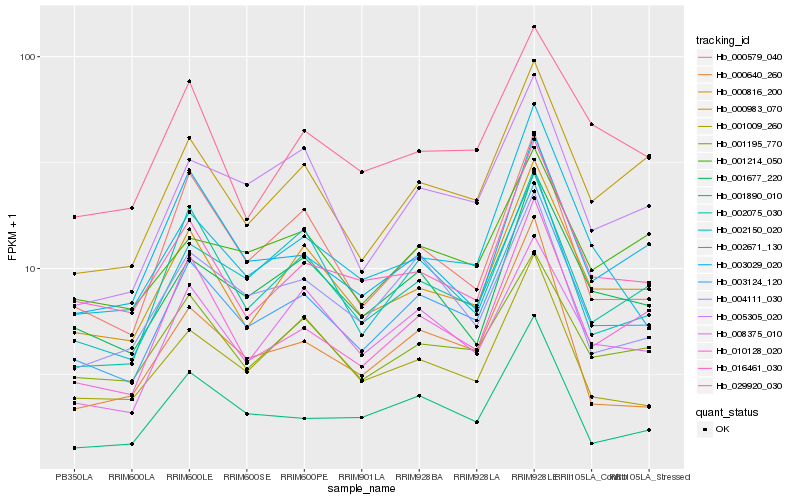
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_120 |
0.0 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 2 |
Hb_000816_200 |
0.069501612 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_010128_020 |
0.0795582689 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 4 |
Hb_016461_030 |
0.0831839452 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_001677_220 |
0.083762051 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 6 |
Hb_000983_070 |
0.0842069319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002075_030 |
0.0916843012 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001890_010 |
0.0921521298 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_001195_770 |
0.0923936175 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 10 |
Hb_005305_020 |
0.0952533119 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000579_040 |
0.0972662015 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 12 |
Hb_002671_130 |
0.0982479518 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 13 |
Hb_008375_010 |
0.0997423421 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 14 |
Hb_002150_020 |
0.101288477 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_029920_030 |
0.1018385372 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001009_260 |
0.1042780623 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000640_260 |
0.1049313242 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004111_030 |
0.1070792173 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_003029_020 |
0.1083307105 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 20 |
Hb_001214_050 |
0.1089753366 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |