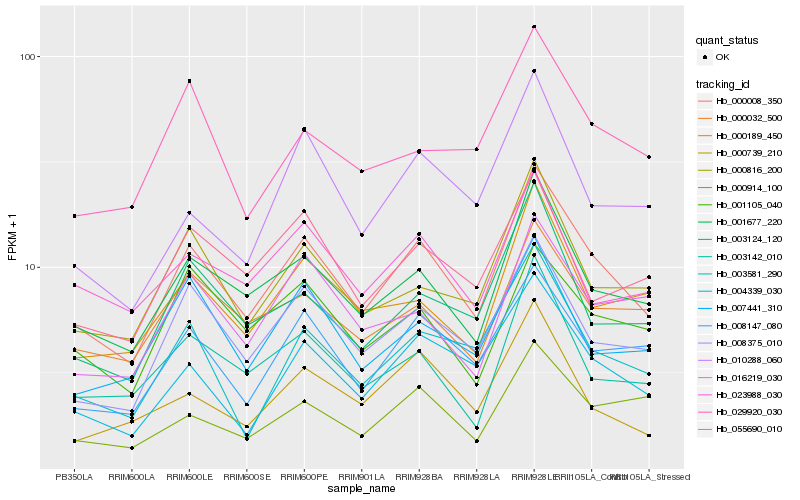
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_350 |
0.0 |
- |
- |
- |
| 2 |
Hb_001677_220 |
0.080611883 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 3 |
Hb_008375_010 |
0.0891740716 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 4 |
Hb_004339_030 |
0.0969368124 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 5 |
Hb_008147_080 |
0.0984917214 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000816_200 |
0.1080427441 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 7 |
Hb_003124_120 |
0.1104725835 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 8 |
Hb_000032_500 |
0.1111125221 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 9 |
Hb_003581_290 |
0.1128498273 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_003142_010 |
0.1132991823 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 11 |
Hb_000189_450 |
0.1144134848 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_007441_310 |
0.1176043581 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_010288_060 |
0.1187337804 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 14 |
Hb_023988_030 |
0.1194162152 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 15 |
Hb_055690_010 |
0.1194523031 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000739_210 |
0.1197447228 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g01970 [Jatropha curcas] |
| 17 |
Hb_001105_040 |
0.1197805584 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000914_100 |
0.1203425416 |
- |
- |
unknown [Glycine max] |
| 19 |
Hb_029920_030 |
0.1223937674 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_016219_030 |
0.1226096184 |
- |
- |
cytochrome P450, putative [Ricinus communis] |