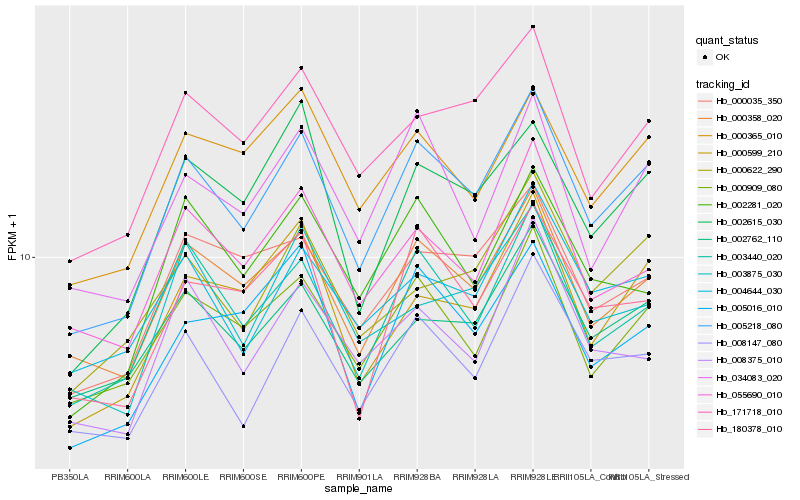
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005218_080 |
0.0 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000358_020 |
0.0689883538 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 3 |
Hb_003440_020 |
0.0789463998 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000909_080 |
0.0811948029 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 5 |
Hb_008147_080 |
0.0815672158 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 6 |
Hb_171718_010 |
0.0872531956 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 7 |
Hb_002762_110 |
0.0896912519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_034083_020 |
0.0927226504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004644_030 |
0.0947984941 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 10 |
Hb_000035_350 |
0.0948378098 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002615_030 |
0.0949536552 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 12 |
Hb_000365_010 |
0.0953821141 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 13 |
Hb_000599_210 |
0.0960809291 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 14 |
Hb_055690_010 |
0.0961060751 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_180378_010 |
0.096978525 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 16 |
Hb_000622_290 |
0.0991215754 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 17 |
Hb_002281_020 |
0.1022677037 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 18 |
Hb_005016_010 |
0.1031606872 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 19 |
Hb_003875_030 |
0.1064656742 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 20 |
Hb_008375_010 |
0.1082951123 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |