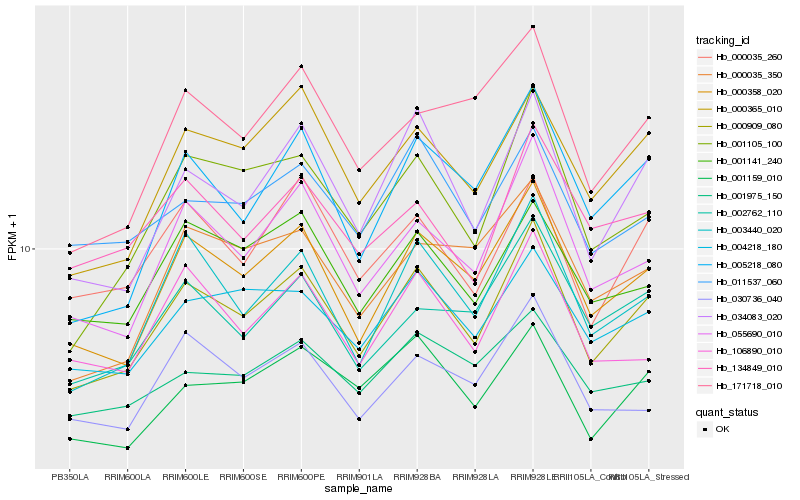
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000909_080 |
0.0 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 2 |
Hb_000358_020 |
0.0575088062 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 3 |
Hb_034083_020 |
0.0664845644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003440_020 |
0.0718762166 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_055690_010 |
0.0730912117 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000365_010 |
0.0788940827 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 7 |
Hb_001105_100 |
0.0797662178 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 8 |
Hb_005218_080 |
0.0811948029 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_004218_180 |
0.0874260037 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 10 |
Hb_000035_260 |
0.0902748403 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 11 |
Hb_001159_010 |
0.0917889619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001141_240 |
0.0928519063 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002762_110 |
0.0950569678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_106890_010 |
0.0954572832 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_011537_060 |
0.0984719401 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 16 |
Hb_134849_010 |
0.0995872348 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_171718_010 |
0.0996387374 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 18 |
Hb_000035_350 |
0.0996531961 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_030736_040 |
0.0998393637 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001975_150 |
0.1009405863 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |