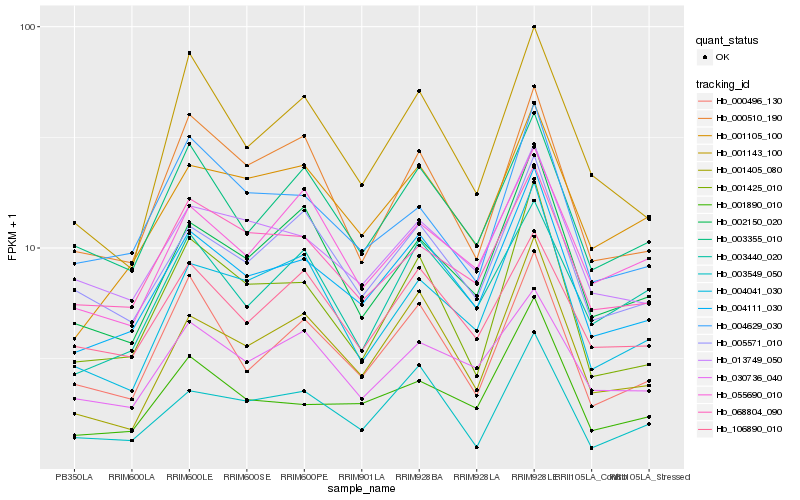
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004111_030 |
0.0 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_013749_050 |
0.0813945208 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005571_010 |
0.0824915488 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 4 |
Hb_001890_010 |
0.0860681111 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_001105_100 |
0.0866087531 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 6 |
Hb_002150_020 |
0.0873547503 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_003355_010 |
0.0884629278 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 8 |
Hb_068804_090 |
0.0898166773 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 9 |
Hb_030736_040 |
0.0906390281 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 10 |
Hb_106890_010 |
0.0931365976 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_001405_080 |
0.0938000952 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 12 |
Hb_001425_010 |
0.0981281195 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 13 |
Hb_000496_130 |
0.099674713 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 14 |
Hb_004629_030 |
0.1002829177 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_004041_030 |
0.1008591608 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 16 |
Hb_055690_010 |
0.1021070795 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001143_100 |
0.1039662981 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 18 |
Hb_000510_190 |
0.104798595 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 19 |
Hb_003440_020 |
0.1048253141 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_003549_050 |
0.1048604505 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |