| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
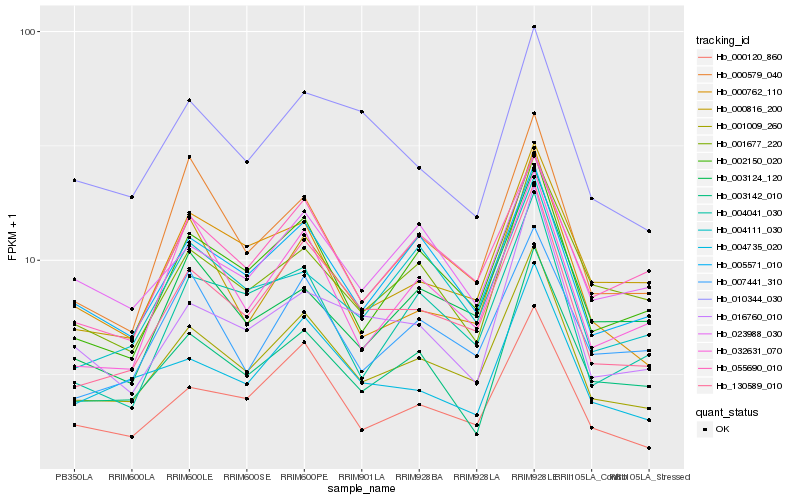
Hb_001009_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_130589_010 |
0.0579682477 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 3 |
Hb_000120_860 |
0.0716910466 |
- |
- |
nucellin, putative [Ricinus communis] |
| 4 |
Hb_002150_020 |
0.0725602575 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_005571_010 |
0.0849154404 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 6 |
Hb_004735_020 |
0.0861485537 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 7 |
Hb_000579_040 |
0.0948379723 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 8 |
Hb_003142_010 |
0.0951950026 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 9 |
Hb_016760_010 |
0.0971665726 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_001677_220 |
0.0977163064 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 11 |
Hb_032631_070 |
0.0978656198 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_007441_310 |
0.0989273886 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000816_200 |
0.1000603165 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 14 |
Hb_055690_010 |
0.1000966673 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004041_030 |
0.1016318873 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 16 |
Hb_023988_030 |
0.1036857312 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 17 |
Hb_003124_120 |
0.1042780623 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 18 |
Hb_000762_110 |
0.1042860674 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004111_030 |
0.1049823398 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_010344_030 |
0.1062066692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |