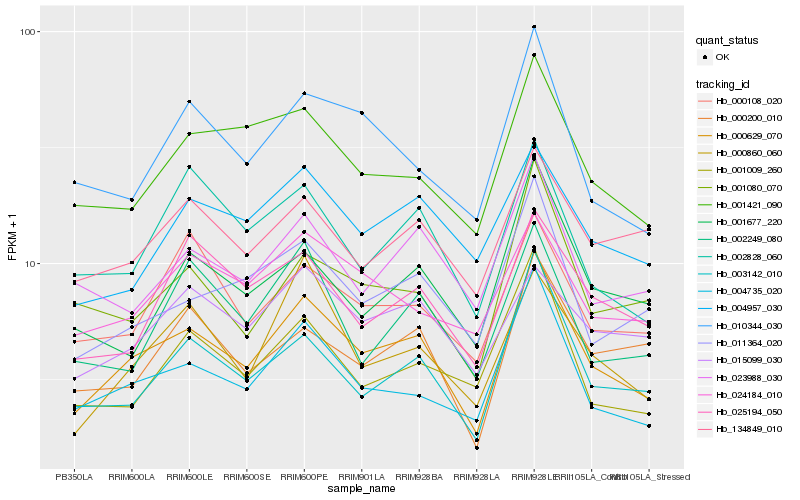
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000629_070 |
0.0 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 2 |
Hb_004735_020 |
0.0980523268 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 3 |
Hb_003142_010 |
0.1035302788 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 4 |
Hb_011364_020 |
0.1134414201 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001421_090 |
0.1156260329 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 6 |
Hb_000860_060 |
0.1174089655 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 7 |
Hb_010344_030 |
0.1195895503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002828_060 |
0.1206622257 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004957_030 |
0.1229646811 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 10 |
Hb_023988_030 |
0.1230667283 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 11 |
Hb_025194_050 |
0.124168835 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001080_070 |
0.1243804524 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 13 |
Hb_000200_010 |
0.1244451461 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 14 |
Hb_000108_020 |
0.1252518317 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 15 |
Hb_024184_010 |
0.1262881125 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_015099_030 |
0.1268258918 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 17 |
Hb_001009_260 |
0.1268779498 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_134849_010 |
0.1271195696 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 19 |
Hb_002249_080 |
0.1275863181 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 20 |
Hb_001677_220 |
0.1276431758 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |