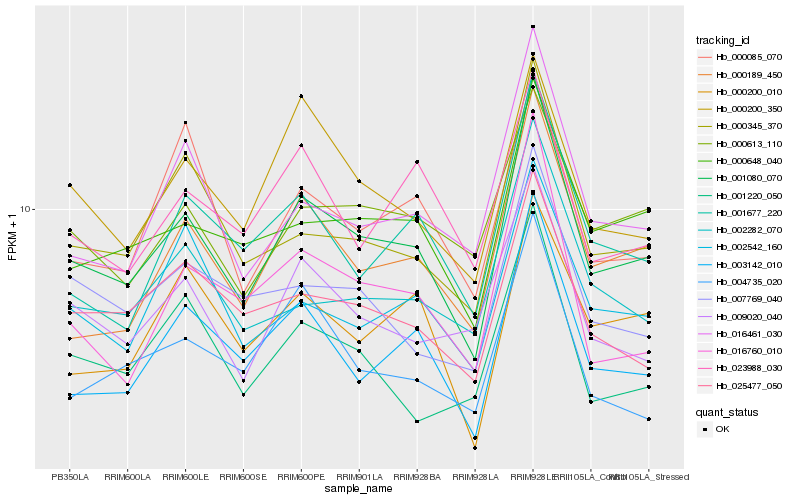
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_070 |
0.0 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 2 |
Hb_000189_450 |
0.085065257 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003142_010 |
0.0880644577 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 4 |
Hb_000345_370 |
0.0939851176 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 5 |
Hb_000648_040 |
0.0955140922 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_016461_030 |
0.098522685 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_001677_220 |
0.1004239231 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 8 |
Hb_000200_350 |
0.10356913 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 9 |
Hb_002282_070 |
0.1042836537 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 10 |
Hb_016760_010 |
0.1043590633 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_025477_050 |
0.1045758843 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_009020_040 |
0.1068910536 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002542_160 |
0.1073380864 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000200_010 |
0.1093922298 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 15 |
Hb_007769_040 |
0.1096924278 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_000613_110 |
0.1101140482 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 17 |
Hb_000085_070 |
0.1144170487 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001220_050 |
0.1145473049 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 19 |
Hb_023988_030 |
0.1148169522 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 20 |
Hb_004735_020 |
0.1149082138 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |