| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
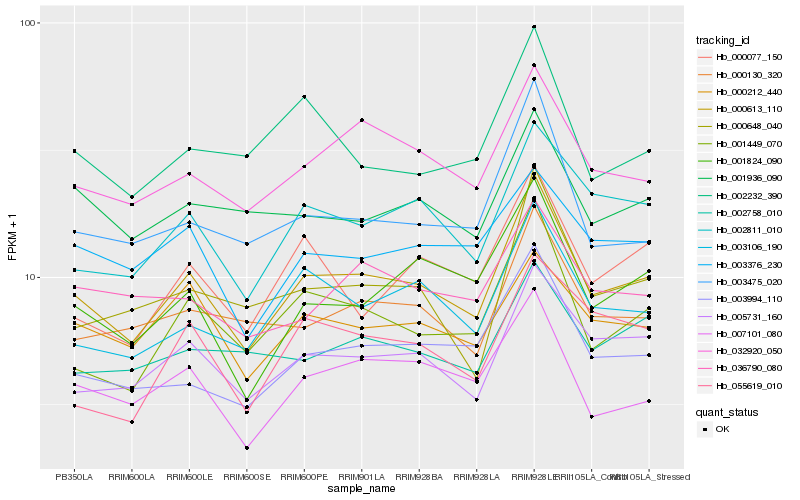
Hb_000613_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 2 |
Hb_005731_160 |
0.0703583266 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_002811_010 |
0.0780668772 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 4 |
Hb_032920_050 |
0.079046257 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001449_070 |
0.0800100676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003106_190 |
0.0821268698 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 7 |
Hb_003994_110 |
0.0826839088 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 8 |
Hb_007101_080 |
0.0846860509 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 9 |
Hb_000130_320 |
0.0890235263 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 10 |
Hb_001824_090 |
0.0910068312 |
- |
- |
- |
| 11 |
Hb_003475_020 |
0.091717138 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 12 |
Hb_000212_440 |
0.0921071787 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_003376_230 |
0.0953003263 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 14 |
Hb_000077_150 |
0.0961330489 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 15 |
Hb_000648_040 |
0.0972459266 |
- |
- |
unknown [Populus trichocarpa] |
| 16 |
Hb_001936_090 |
0.09748919 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_036790_080 |
0.098818679 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 18 |
Hb_002758_010 |
0.1000737419 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 19 |
Hb_002232_390 |
0.1022774753 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 20 |
Hb_055619_010 |
0.102647259 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |