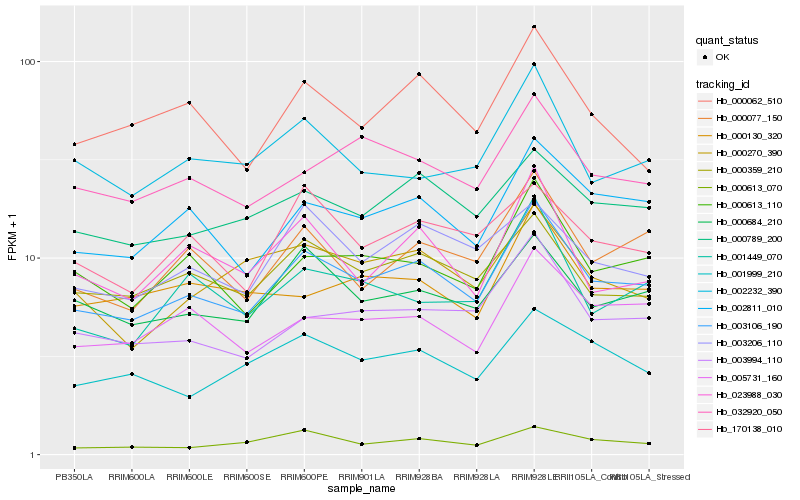
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_190 |
0.0 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 2 |
Hb_002811_010 |
0.0761116356 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 3 |
Hb_000613_110 |
0.0821268698 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 4 |
Hb_000359_210 |
0.0832532509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003994_110 |
0.0841416574 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 6 |
Hb_000684_210 |
0.0844775117 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 7 |
Hb_000077_150 |
0.0849497123 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 8 |
Hb_000789_200 |
0.0871418902 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_032920_050 |
0.0922238584 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005731_160 |
0.0925517505 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000062_510 |
0.0954809113 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_023988_030 |
0.0957026929 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 13 |
Hb_003206_110 |
0.0975354638 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 14 |
Hb_002232_390 |
0.0998185547 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 15 |
Hb_001449_070 |
0.0999395539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_170138_010 |
0.1001552122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000613_070 |
0.1010396651 |
- |
- |
- |
| 18 |
Hb_001999_210 |
0.1013579965 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 19 |
Hb_000270_390 |
0.1017392872 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 20 |
Hb_000130_320 |
0.1021535651 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |