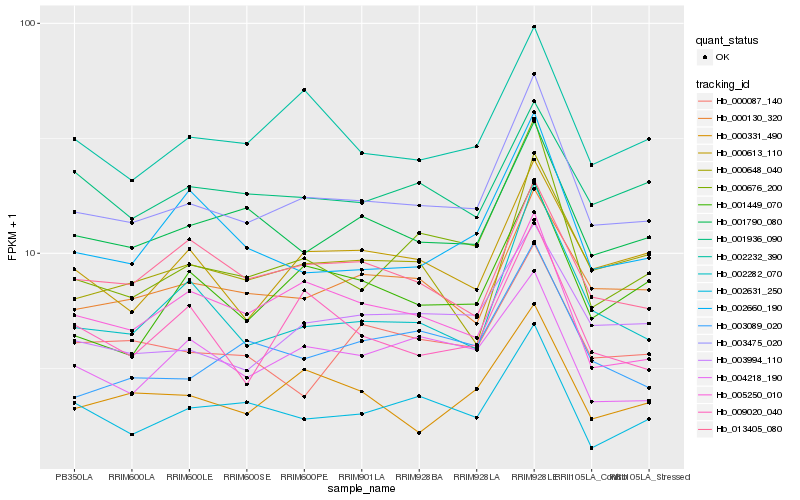
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003475_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 2 |
Hb_003994_110 |
0.0816521116 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 3 |
Hb_001790_080 |
0.0817637104 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 4 |
Hb_000676_200 |
0.0829441967 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 5 |
Hb_000130_320 |
0.0833939203 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 6 |
Hb_002232_390 |
0.0877375562 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 7 |
Hb_002282_070 |
0.0881868221 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 8 |
Hb_000613_110 |
0.091717138 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 9 |
Hb_001449_070 |
0.0981493488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002631_250 |
0.0982923464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 11 |
Hb_000331_490 |
0.0983164053 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005250_010 |
0.0985160204 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 13 |
Hb_013405_080 |
0.1001476048 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 14 |
Hb_000648_040 |
0.1007384983 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_000087_140 |
0.1011316412 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002660_190 |
0.1025390956 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 17 |
Hb_003089_020 |
0.103990343 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g18940, chloroplastic [Jatropha curcas] |
| 18 |
Hb_009020_040 |
0.1044247523 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004218_190 |
0.1046162841 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001936_090 |
0.1056089321 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |