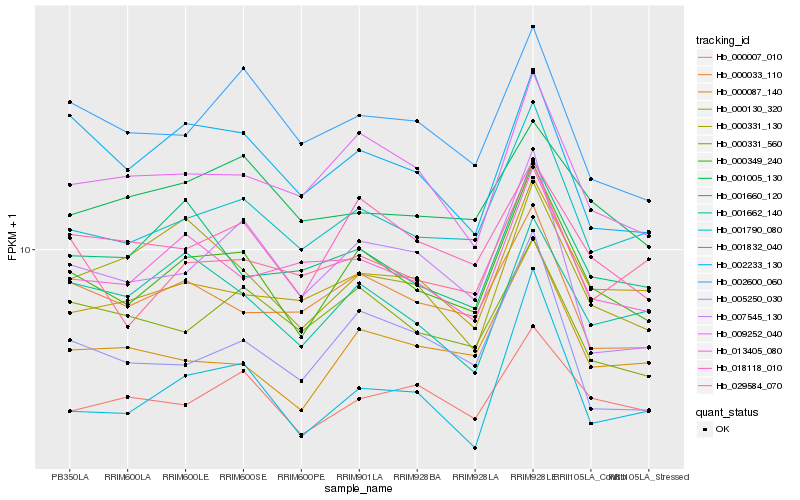
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000349_240 |
0.0 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001790_080 |
0.0704672566 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 3 |
Hb_018118_010 |
0.0844536757 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 4 |
Hb_009252_040 |
0.0888056045 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000087_140 |
0.0950946612 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000033_110 |
0.097140305 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 7 |
Hb_000007_010 |
0.0974522493 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000130_320 |
0.0990371345 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 9 |
Hb_013405_080 |
0.1020898274 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 10 |
Hb_000331_560 |
0.1038901658 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 11 |
Hb_001662_140 |
0.1039617939 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002600_060 |
0.1050004755 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 13 |
Hb_029584_070 |
0.1070524428 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 14 |
Hb_002233_130 |
0.1083062801 |
- |
- |
transporter, putative [Ricinus communis] |
| 15 |
Hb_007545_130 |
0.1085482196 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005250_030 |
0.108840822 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 17 |
Hb_000331_130 |
0.1090686977 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001660_120 |
0.1103411532 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001005_130 |
0.1107516126 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 20 |
Hb_001832_040 |
0.1112113728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |