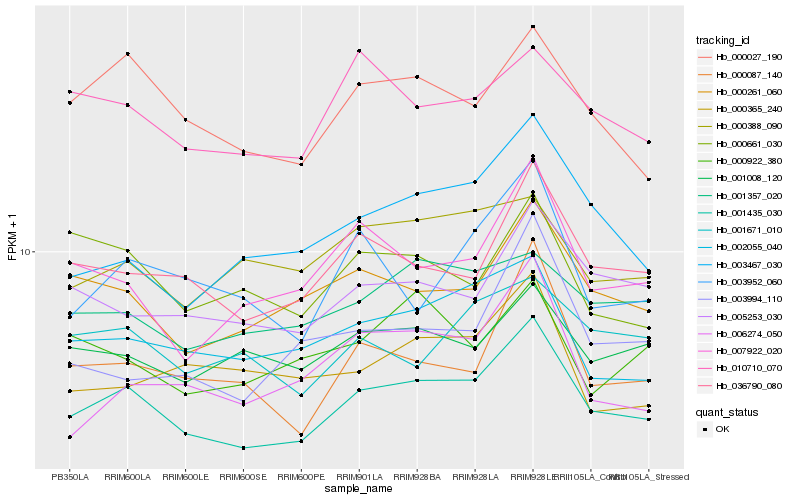
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001435_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 2 |
Hb_036790_080 |
0.0836959382 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 3 |
Hb_000261_060 |
0.0868404588 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 4 |
Hb_000027_190 |
0.0893221329 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002055_040 |
0.0910465621 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 6 |
Hb_005253_030 |
0.0949004087 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_003467_030 |
0.0952074477 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_007922_020 |
0.0956242129 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 9 |
Hb_000087_140 |
0.0993944766 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001357_020 |
0.1006638411 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001008_120 |
0.1028406086 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 12 |
Hb_010710_070 |
0.104681209 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_003952_060 |
0.1065184091 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 14 |
Hb_000388_090 |
0.1065765192 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_003994_110 |
0.1070434982 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 16 |
Hb_001671_010 |
0.1087993502 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 17 |
Hb_006274_050 |
0.1097774695 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 18 |
Hb_000661_030 |
0.111435824 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 19 |
Hb_000365_240 |
0.1124906502 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 20 |
Hb_000922_380 |
0.1130077456 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein MPP10 isoform X1 [Jatropha curcas] |