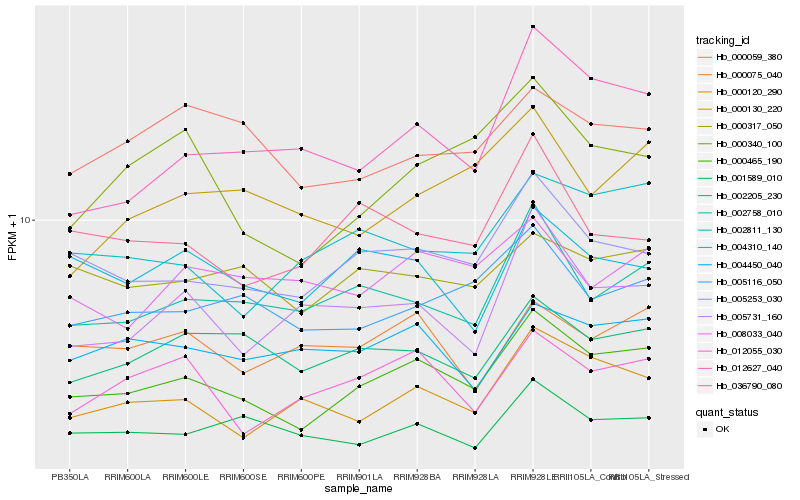
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_190 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005253_030 |
0.081331726 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_002758_010 |
0.0975063921 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 4 |
Hb_005116_050 |
0.0989273045 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000120_290 |
0.1019074352 |
- |
- |
PREDICTED: N-acylphosphatidylethanolamine synthase isoform X1 [Jatropha curcas] |
| 6 |
Hb_002205_230 |
0.1032918355 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000340_100 |
0.1036151023 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002811_130 |
0.1038754458 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_012627_040 |
0.1060061512 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 10 |
Hb_001589_010 |
0.1079894384 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000317_050 |
0.1081627906 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 12 |
Hb_012055_030 |
0.1091298015 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 13 |
Hb_004450_040 |
0.1110538132 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 14 |
Hb_005731_160 |
0.1124358001 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_036790_080 |
0.1131389251 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 16 |
Hb_004310_140 |
0.1152570896 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_008033_040 |
0.1168648384 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000130_220 |
0.1171662697 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000075_040 |
0.1179469594 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 20 |
Hb_000059_380 |
0.1182568285 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |