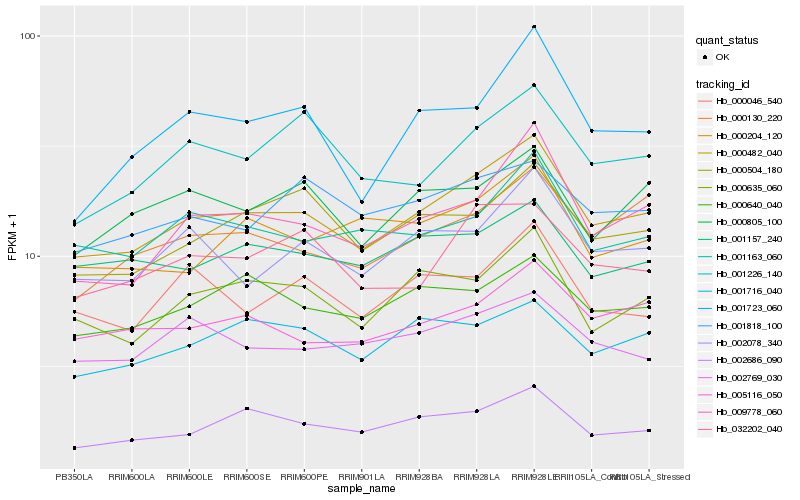
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000482_040 |
0.0 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 2 |
Hb_009778_060 |
0.0675581058 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 3 |
Hb_002686_090 |
0.0754005412 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 4 |
Hb_000635_060 |
0.0755176836 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 5 |
Hb_002078_340 |
0.0788563256 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005116_050 |
0.079929899 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_001163_060 |
0.0819076586 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 8 |
Hb_000130_220 |
0.0821219361 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_032202_040 |
0.0848736365 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 10 |
Hb_000504_180 |
0.0863235942 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 11 |
Hb_001723_060 |
0.0890439185 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 12 |
Hb_000640_040 |
0.0891811273 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000204_120 |
0.089294051 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 14 |
Hb_001226_140 |
0.0898323574 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 15 |
Hb_001157_240 |
0.0906032525 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 16 |
Hb_001716_040 |
0.0909545939 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 17 |
Hb_000046_540 |
0.0911562503 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 18 |
Hb_002769_030 |
0.0912947549 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000805_100 |
0.0913980322 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_001818_100 |
0.0915059309 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |