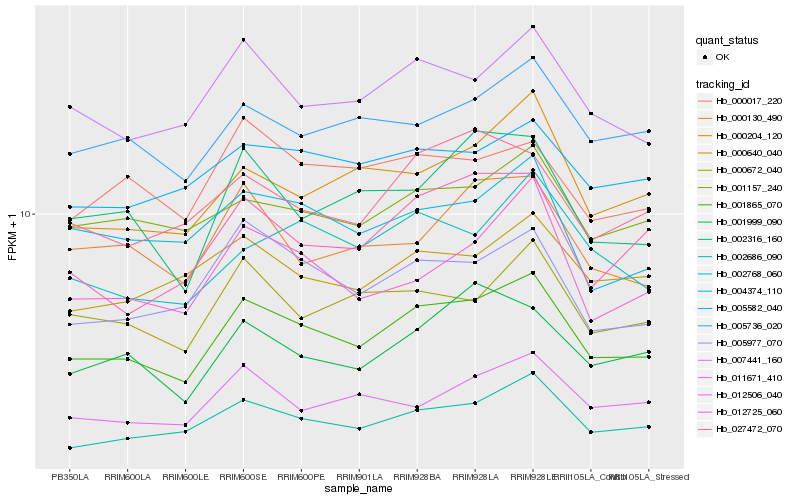
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001865_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002686_090 |
0.0654555002 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 3 |
Hb_007441_160 |
0.0701248564 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_005977_070 |
0.0744280455 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000672_040 |
0.0785339824 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 6 |
Hb_005736_020 |
0.0792666824 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 7 |
Hb_001157_240 |
0.0795981921 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 8 |
Hb_000204_120 |
0.0801128638 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 9 |
Hb_012506_040 |
0.0804965275 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 10 |
Hb_002316_160 |
0.0812971288 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 11 |
Hb_004374_110 |
0.0815050689 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 12 |
Hb_012725_060 |
0.0815832057 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_005582_040 |
0.083585111 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 14 |
Hb_001999_090 |
0.0851310656 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002768_060 |
0.0853212695 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000640_040 |
0.0856338513 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_027472_070 |
0.0856908099 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 18 |
Hb_011671_410 |
0.0862245544 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 19 |
Hb_000130_490 |
0.0869643618 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000017_220 |
0.0879521279 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |