| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
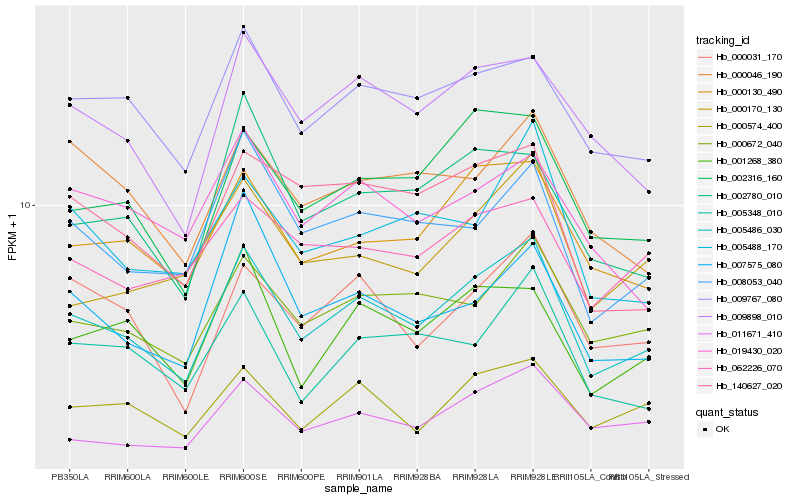
Hb_005486_030 |
0.0 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 2 |
Hb_000130_490 |
0.0707372812 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000672_040 |
0.0724535605 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 4 |
Hb_002316_160 |
0.0726837259 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 5 |
Hb_009767_080 |
0.0750210337 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 6 |
Hb_009898_010 |
0.0786642773 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 7 |
Hb_008053_040 |
0.0801583748 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 8 |
Hb_011671_410 |
0.0817310876 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 9 |
Hb_000574_400 |
0.0819445749 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 10 |
Hb_001268_380 |
0.0825231975 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 11 |
Hb_000031_170 |
0.0830826579 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005348_010 |
0.0864632125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002780_010 |
0.0872406742 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 14 |
Hb_005488_170 |
0.0904028364 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 15 |
Hb_140627_020 |
0.0906123365 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000170_130 |
0.0908474878 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 17 |
Hb_062226_070 |
0.0919148611 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_190 |
0.0927788247 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 19 |
Hb_007575_080 |
0.0928814945 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 20 |
Hb_019430_020 |
0.0958144011 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |