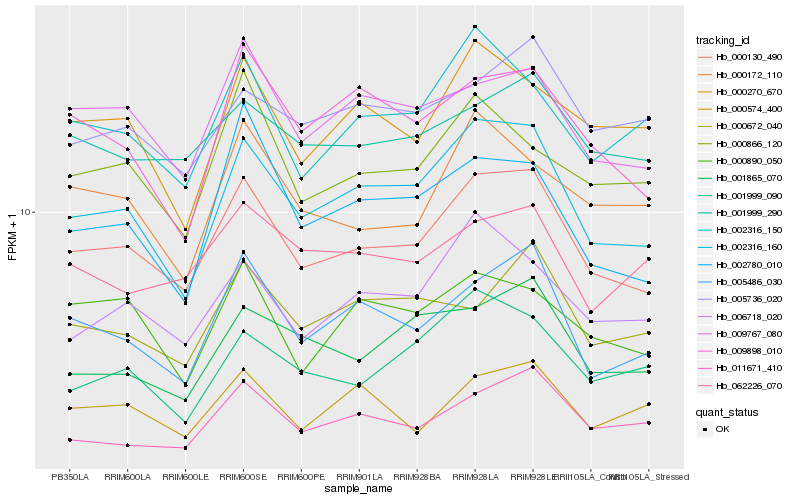
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_490 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002316_160 |
0.0526726601 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 3 |
Hb_005486_030 |
0.0707372812 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 4 |
Hb_009767_080 |
0.0731236664 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 5 |
Hb_006718_020 |
0.0792460606 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 6 |
Hb_011671_410 |
0.081144838 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 7 |
Hb_001999_290 |
0.0815133127 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002316_150 |
0.0862388078 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 9 |
Hb_001865_070 |
0.0869643618 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009898_010 |
0.0880026571 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 11 |
Hb_002780_010 |
0.0888001475 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 12 |
Hb_000866_120 |
0.0893904798 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 13 |
Hb_000890_050 |
0.0895904567 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 14 |
Hb_001999_090 |
0.0902247383 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000270_670 |
0.0903835872 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 16 |
Hb_000172_110 |
0.0905438354 |
- |
- |
- |
| 17 |
Hb_000574_400 |
0.0908196742 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 18 |
Hb_005736_020 |
0.091281645 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 19 |
Hb_000672_040 |
0.0923227357 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 20 |
Hb_062226_070 |
0.0924353528 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |