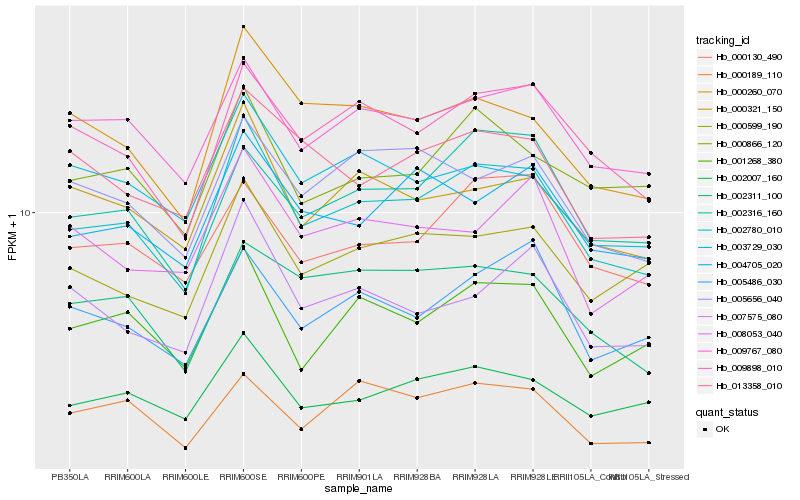
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002780_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 2 |
Hb_002007_160 |
0.0737927708 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 3 |
Hb_009898_010 |
0.0738725417 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 4 |
Hb_009767_080 |
0.0764093026 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 5 |
Hb_000321_150 |
0.0800526387 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000260_070 |
0.0802224123 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_001268_380 |
0.080468305 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 8 |
Hb_004705_020 |
0.0835558081 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 9 |
Hb_000599_190 |
0.0835772044 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 10 |
Hb_002316_160 |
0.0851410002 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 11 |
Hb_000866_120 |
0.0868034214 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 12 |
Hb_005486_030 |
0.0872406742 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 13 |
Hb_000130_490 |
0.0888001475 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002311_100 |
0.0903281323 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 15 |
Hb_007575_080 |
0.0903939926 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 16 |
Hb_005656_040 |
0.0907745555 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 17 |
Hb_013358_010 |
0.0913449774 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 18 |
Hb_003729_030 |
0.0952907468 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 19 |
Hb_008053_040 |
0.0969153717 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 20 |
Hb_000189_110 |
0.098553743 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04750, mitochondrial [Jatropha curcas] |