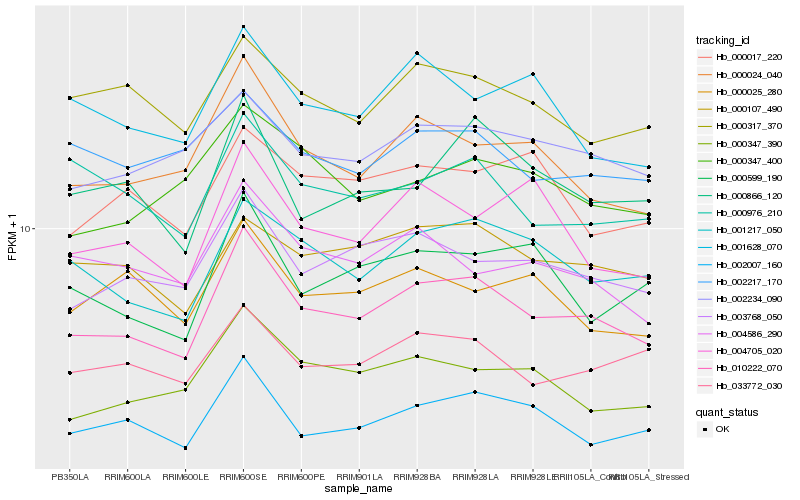
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010222_070 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_003768_050 |
0.0724028493 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000024_040 |
0.0725390027 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 4 |
Hb_001217_050 |
0.0743532833 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 5 |
Hb_002007_160 |
0.0747371056 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 6 |
Hb_000347_390 |
0.0816838917 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 7 |
Hb_002234_090 |
0.0832817363 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 8 |
Hb_033772_030 |
0.0835816268 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 9 |
Hb_000107_490 |
0.0842110179 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 10 |
Hb_000866_120 |
0.0857251982 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 11 |
Hb_000599_190 |
0.0867946226 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 12 |
Hb_004586_290 |
0.0873855918 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 13 |
Hb_000317_370 |
0.089138475 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 14 |
Hb_004705_020 |
0.0903013463 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 15 |
Hb_000976_210 |
0.0903410052 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000347_400 |
0.0919524296 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001628_070 |
0.0926765902 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_000025_280 |
0.0931433912 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 19 |
Hb_000017_220 |
0.0950886936 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 20 |
Hb_002217_170 |
0.0953450469 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |