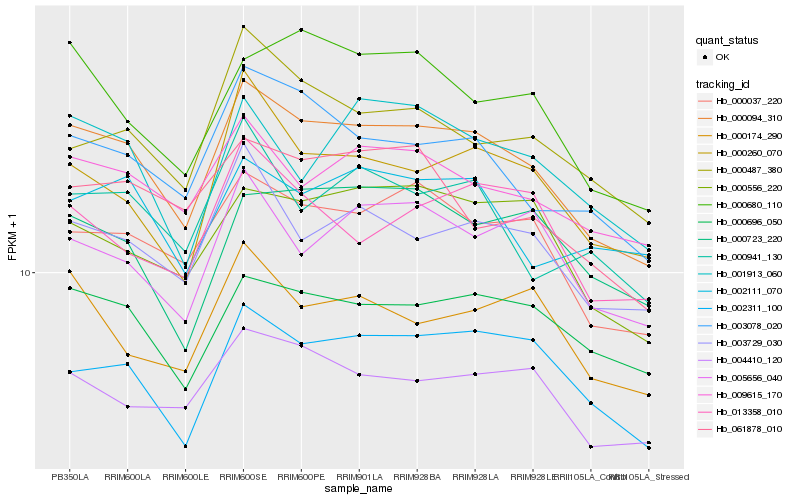
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 2 |
Hb_000037_220 |
0.0679858993 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 3 |
Hb_000696_050 |
0.0722272178 |
- |
- |
PREDICTED: CSC1-like protein At4g35870 [Jatropha curcas] |
| 4 |
Hb_003078_020 |
0.0724579289 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 5 |
Hb_003729_030 |
0.0750869802 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 6 |
Hb_002311_100 |
0.0757266 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 7 |
Hb_009615_170 |
0.0775023376 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004410_120 |
0.0797763014 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_070 |
0.081182035 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000556_220 |
0.0812748249 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 11 |
Hb_000487_380 |
0.0868504906 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000941_130 |
0.0872375417 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 13 |
Hb_000680_110 |
0.0876900151 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 14 |
Hb_005656_040 |
0.0883337025 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_001913_060 |
0.0888922729 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 16 |
Hb_061878_010 |
0.0900282462 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 17 |
Hb_000723_220 |
0.0904701591 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 18 |
Hb_002111_070 |
0.0911043758 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 19 |
Hb_000174_290 |
0.0921749008 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 20 |
Hb_013358_010 |
0.0923480775 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |