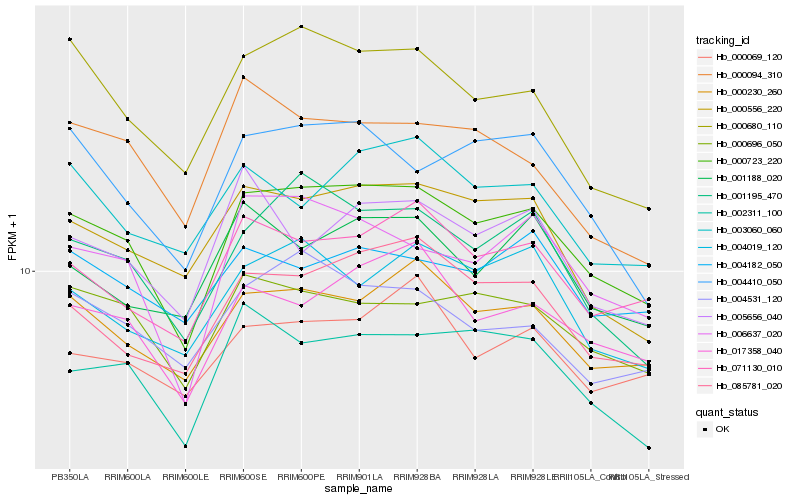
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 2 |
Hb_000230_260 |
0.067156528 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 3 |
Hb_006637_020 |
0.0680412754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 4 |
Hb_017358_040 |
0.06905095 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 5 |
Hb_001195_470 |
0.0705426529 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 6 |
Hb_085781_020 |
0.0726784902 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 7 |
Hb_071130_010 |
0.0733332523 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002311_100 |
0.0733462723 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 9 |
Hb_000696_050 |
0.0734197132 |
- |
- |
PREDICTED: CSC1-like protein At4g35870 [Jatropha curcas] |
| 10 |
Hb_000556_220 |
0.0770294199 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 11 |
Hb_000680_110 |
0.0801028251 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 12 |
Hb_004410_050 |
0.0815490245 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 13 |
Hb_004019_120 |
0.0824727809 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_004531_120 |
0.0826483919 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 15 |
Hb_001188_020 |
0.087344336 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000069_120 |
0.0891095999 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 17 |
Hb_003060_060 |
0.0891372924 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 18 |
Hb_005656_040 |
0.089834759 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000094_310 |
0.0904701591 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 20 |
Hb_004182_050 |
0.0904806874 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |