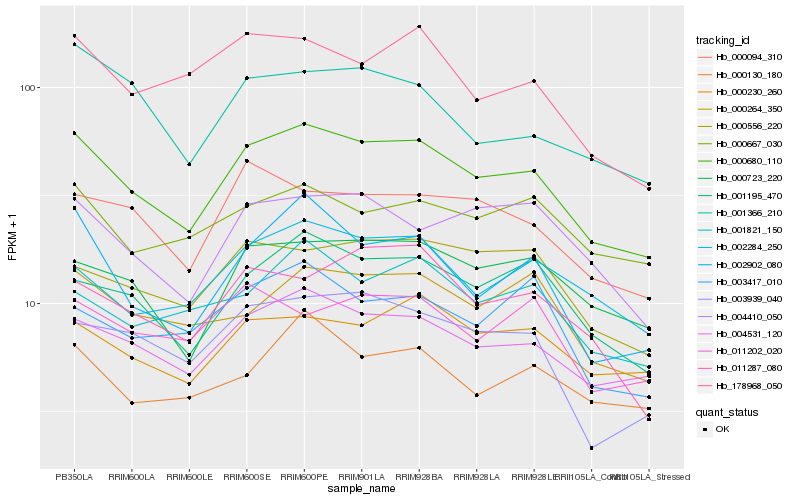
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_110 |
0.0 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 2 |
Hb_004531_120 |
0.0600622661 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 3 |
Hb_001195_470 |
0.0757848215 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 4 |
Hb_000723_220 |
0.0801028251 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 5 |
Hb_000264_350 |
0.0856188507 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 6 |
Hb_000230_260 |
0.0860963049 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 7 |
Hb_002902_080 |
0.0862355961 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 8 |
Hb_000556_220 |
0.0871047656 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 9 |
Hb_000094_310 |
0.0876900151 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 10 |
Hb_002284_250 |
0.0891909579 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 11 |
Hb_004410_050 |
0.0903226262 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 12 |
Hb_003939_040 |
0.091782674 |
- |
- |
hypothetical protein CICLE_v10014350mg [Citrus clementina] |
| 13 |
Hb_001821_150 |
0.0922827327 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 14 |
Hb_178968_050 |
0.0936687374 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 15 |
Hb_000667_030 |
0.094372771 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 16 |
Hb_000130_180 |
0.0957423706 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_011287_080 |
0.0960323152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001366_210 |
0.0964461306 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 19 |
Hb_003417_010 |
0.0980087432 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 20 |
Hb_011202_020 |
0.0985255383 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |