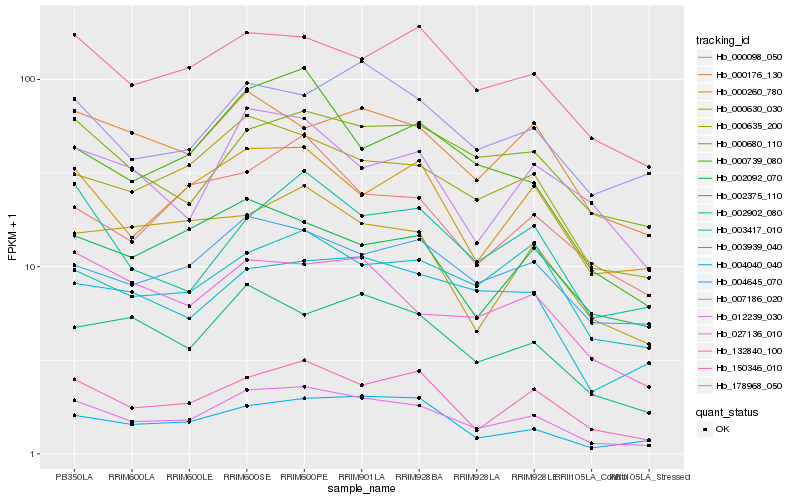
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027136_010 |
0.0 |
- |
- |
hypothetical protein L484_010426 [Morus notabilis] |
| 2 |
Hb_150346_010 |
0.0889706477 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_000635_200 |
0.0952601888 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 4 |
Hb_002375_110 |
0.1083527847 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_178968_050 |
0.1096966848 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 6 |
Hb_004040_040 |
0.1109351105 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 7 |
Hb_000260_780 |
0.1109484473 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |
| 8 |
Hb_003939_040 |
0.1116134858 |
- |
- |
hypothetical protein CICLE_v10014350mg [Citrus clementina] |
| 9 |
Hb_132840_100 |
0.1129355387 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 10 |
Hb_000176_130 |
0.1147762524 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 11 |
Hb_004645_070 |
0.1179974716 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 12 |
Hb_000739_080 |
0.1187704844 |
- |
- |
PREDICTED: 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase [Jatropha curcas] |
| 13 |
Hb_002902_080 |
0.1212651672 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 14 |
Hb_000680_110 |
0.1214843963 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 15 |
Hb_002092_070 |
0.1245868028 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 16 |
Hb_007186_020 |
0.1248805505 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 17 |
Hb_000630_030 |
0.1262661529 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 18 |
Hb_000098_050 |
0.1265339639 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 19 |
Hb_003417_010 |
0.127015686 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 20 |
Hb_012239_030 |
0.1289222303 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |