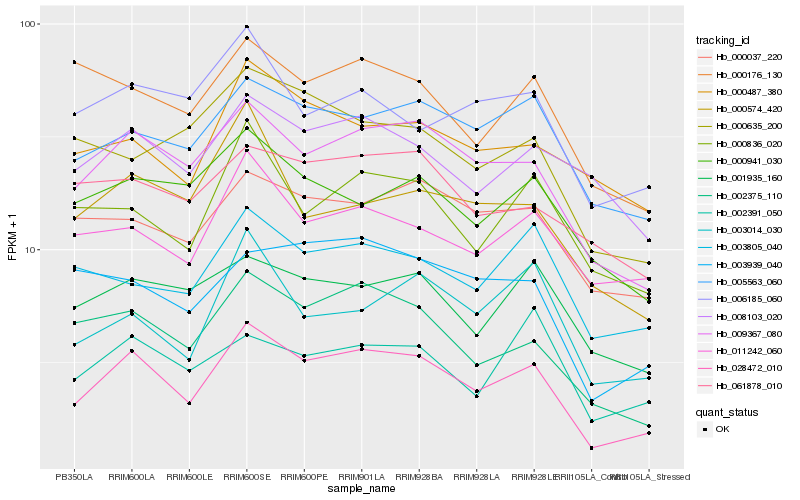
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028472_010 |
0.0 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_009367_080 |
0.0844073013 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 3 |
Hb_002375_110 |
0.1033223332 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_005563_060 |
0.1130851185 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 5 |
Hb_000836_020 |
0.120821547 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 6 |
Hb_000037_220 |
0.123791528 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 7 |
Hb_011242_060 |
0.1258696597 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 8 |
Hb_003939_040 |
0.126830807 |
- |
- |
hypothetical protein CICLE_v10014350mg [Citrus clementina] |
| 9 |
Hb_003014_030 |
0.1272645842 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |
| 10 |
Hb_001935_160 |
0.1285936271 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 11 |
Hb_003805_040 |
0.1301864746 |
- |
- |
Sacsin [Glycine soja] |
| 12 |
Hb_000941_030 |
0.1309175577 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 13 |
Hb_008103_020 |
0.131402821 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 14 |
Hb_000487_380 |
0.1336820837 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000176_130 |
0.1342307781 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 16 |
Hb_002391_050 |
0.135373866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 17 |
Hb_061878_010 |
0.1353908665 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 18 |
Hb_006185_060 |
0.1355257173 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 19 |
Hb_000574_420 |
0.1365960308 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 20 |
Hb_000635_200 |
0.137717815 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |