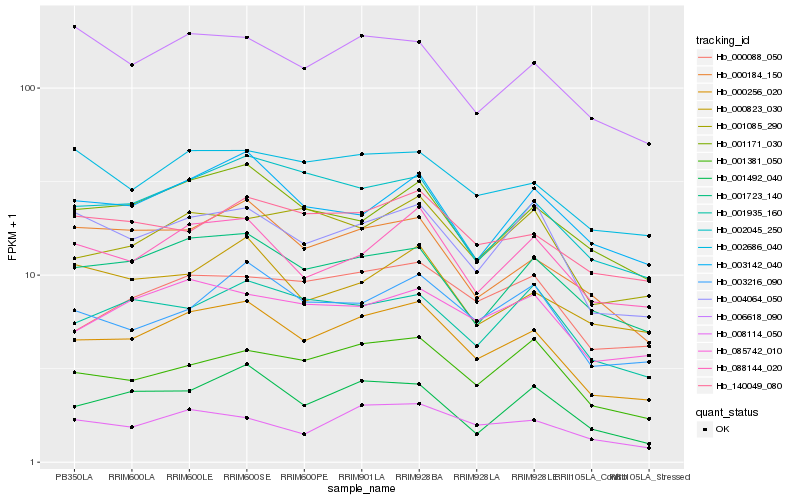
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_020 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 2 |
Hb_001723_140 |
0.0772711221 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_088144_020 |
0.0802193601 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004064_050 |
0.0829269401 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 5 |
Hb_000184_150 |
0.0851704876 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 6 |
Hb_002045_250 |
0.0870811601 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 7 |
Hb_001171_030 |
0.0875938266 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 8 |
Hb_003216_090 |
0.089541677 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008114_050 |
0.0902896541 |
- |
- |
hypothetical protein POPTR_0006s02430g [Populus trichocarpa] |
| 10 |
Hb_001381_050 |
0.0914818207 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 11 |
Hb_001492_040 |
0.0926444896 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000088_050 |
0.0932989254 |
- |
- |
PREDICTED: threonylcarbamoyladenosine tRNA methylthiotransferase [Jatropha curcas] |
| 13 |
Hb_002686_040 |
0.0938348837 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 14 |
Hb_085742_010 |
0.0942697524 |
- |
- |
hypothetical protein POPTR_0005s05170g [Populus trichocarpa] |
| 15 |
Hb_003142_040 |
0.095112781 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 16 |
Hb_001085_290 |
0.0967255009 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 17 |
Hb_006618_090 |
0.0969886186 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 18 |
Hb_001935_160 |
0.0970544782 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 19 |
Hb_140049_080 |
0.0970698607 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 20 |
Hb_000823_030 |
0.0972040033 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |