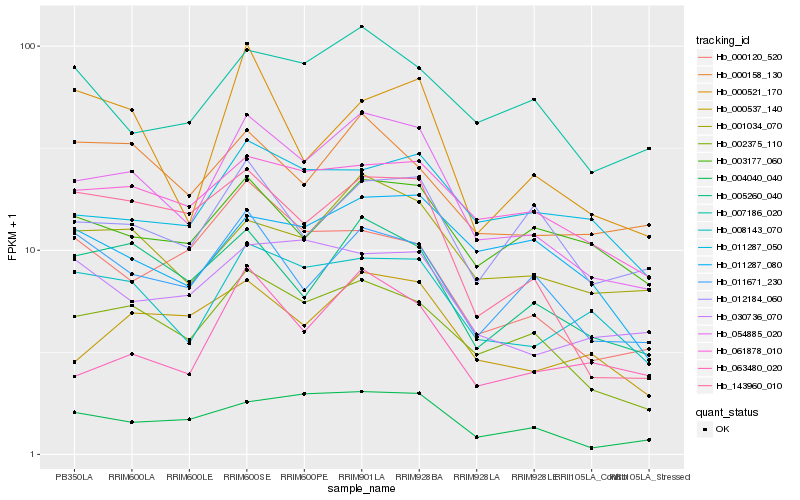
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054885_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002375_110 |
0.0930932616 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_001034_070 |
0.1166843206 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_008143_070 |
0.1178380538 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 5 |
Hb_005260_040 |
0.1195759891 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004040_040 |
0.120011358 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 7 |
Hb_000537_140 |
0.1242809745 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 8 |
Hb_063480_020 |
0.1317122401 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_061878_010 |
0.1331569015 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 10 |
Hb_143960_010 |
0.1344894088 |
- |
- |
- |
| 11 |
Hb_011671_230 |
0.1356695867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007186_020 |
0.1393712039 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 13 |
Hb_012184_060 |
0.1405934206 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000158_130 |
0.140782168 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 15 |
Hb_030736_070 |
0.1426656151 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_011287_050 |
0.1441058277 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 17 |
Hb_000120_520 |
0.145550699 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 18 |
Hb_000521_170 |
0.146191727 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 19 |
Hb_003177_060 |
0.1462611887 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011287_080 |
0.1467448256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |