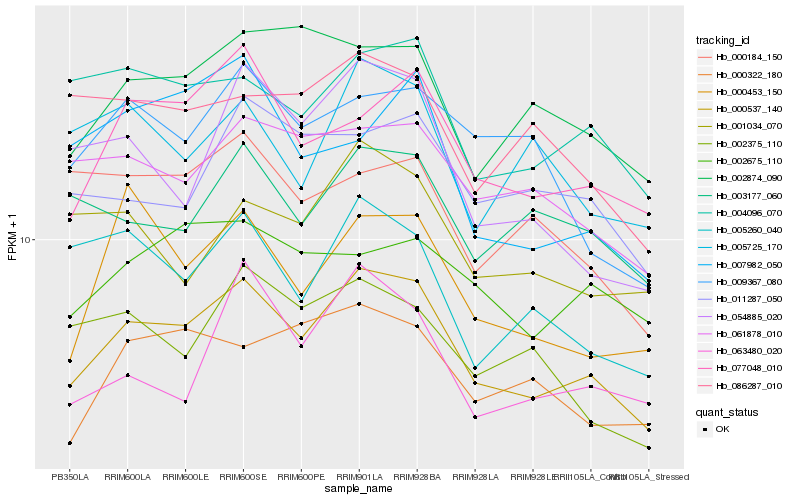
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000537_140 |
0.0 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 2 |
Hb_077048_010 |
0.1117734986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_054885_020 |
0.1242809745 |
- |
- |
- |
| 4 |
Hb_004096_070 |
0.128022207 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000453_150 |
0.1292820912 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_002874_090 |
0.1307852642 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 7 |
Hb_061878_010 |
0.1318445306 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 8 |
Hb_005260_040 |
0.1348508939 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003177_060 |
0.1366410731 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000322_180 |
0.136848463 |
- |
- |
recA family protein [Populus trichocarpa] |
| 11 |
Hb_011287_050 |
0.1375185384 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 12 |
Hb_009367_080 |
0.1384069037 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 13 |
Hb_063480_020 |
0.1406089599 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_002675_110 |
0.1406534093 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 15 |
Hb_001034_070 |
0.141410227 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_002375_110 |
0.1422881284 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_005725_170 |
0.1423931114 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 18 |
Hb_007982_050 |
0.1441850045 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Jatropha curcas] |
| 19 |
Hb_086287_010 |
0.1442769494 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000184_150 |
0.1448866679 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |