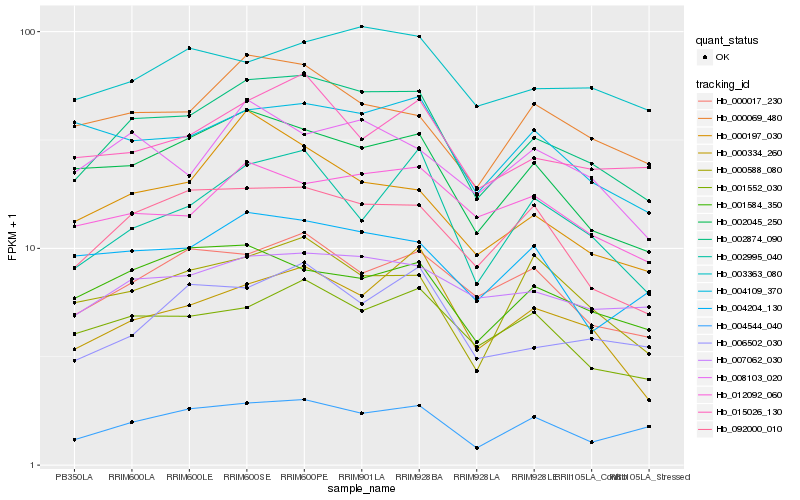
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002874_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 2 |
Hb_002045_250 |
0.0863235524 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 3 |
Hb_092000_010 |
0.0867378333 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 4 |
Hb_004544_040 |
0.0919208767 |
- |
- |
hypothetical protein PHAVU_010G103000g [Phaseolus vulgaris] |
| 5 |
Hb_007062_030 |
0.0940413495 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 6 |
Hb_001552_030 |
0.0945631338 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000017_230 |
0.0958381589 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 8 |
Hb_000069_480 |
0.0958643329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_015026_130 |
0.0959977862 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 10 |
Hb_006502_030 |
0.0981293521 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 11 |
Hb_000334_260 |
0.0992939665 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 12 |
Hb_001584_350 |
0.1006634003 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 13 |
Hb_002995_040 |
0.1010874099 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_012092_060 |
0.1020760963 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 15 |
Hb_003363_080 |
0.1030246885 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 16 |
Hb_000588_080 |
0.1033153205 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 17 |
Hb_008103_020 |
0.104209149 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 18 |
Hb_000197_030 |
0.1045165935 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 19 |
Hb_004109_370 |
0.1047067842 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 20 |
Hb_004204_130 |
0.1049005597 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |