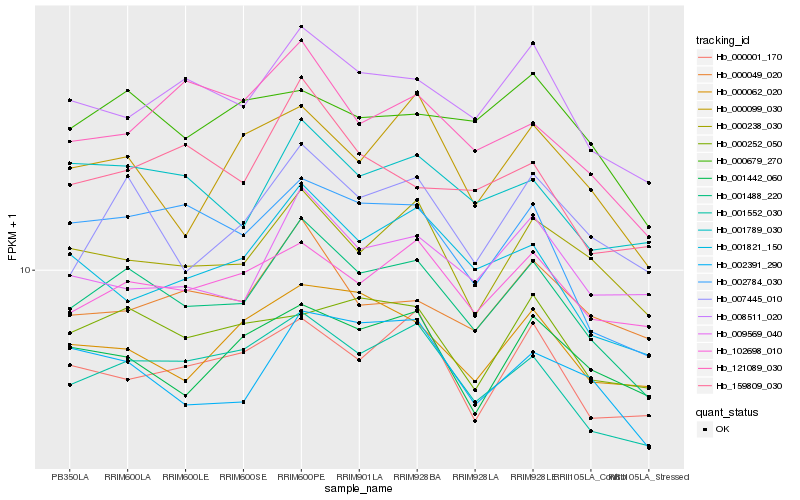
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_220 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 2 |
Hb_001552_030 |
0.0661550383 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_008511_020 |
0.0775516029 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 4 |
Hb_002784_030 |
0.079279449 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 5 |
Hb_007445_010 |
0.0809768543 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_000238_030 |
0.0846222422 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_001789_030 |
0.0876573265 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 8 |
Hb_102698_010 |
0.0887285 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002391_290 |
0.0902244251 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001821_150 |
0.0925719657 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 11 |
Hb_000062_020 |
0.0938495223 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_121089_030 |
0.0948510106 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 13 |
Hb_000001_170 |
0.0953785765 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_000099_030 |
0.0959112753 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000252_050 |
0.0960335343 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 16 |
Hb_159809_030 |
0.0966552483 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000679_270 |
0.0967111946 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 18 |
Hb_000049_020 |
0.0978469347 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 19 |
Hb_001442_060 |
0.09807766 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009569_040 |
0.0995098612 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |