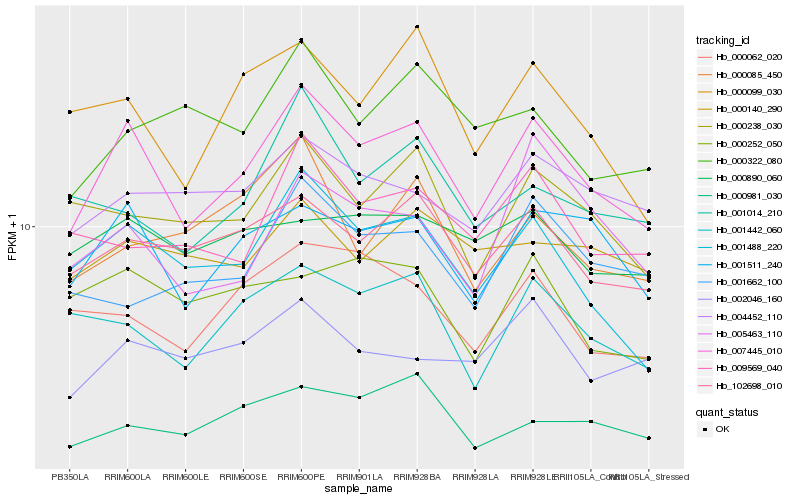
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007445_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_001511_240 |
0.0738798869 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 3 |
Hb_001488_220 |
0.0809768543 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 4 |
Hb_102698_010 |
0.0874834152 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001442_060 |
0.0933346731 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000099_030 |
0.0974234617 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000238_030 |
0.098459798 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000062_020 |
0.1006943451 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_004452_110 |
0.1022285811 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009569_040 |
0.1037074287 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 11 |
Hb_002046_160 |
0.1047996763 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000981_030 |
0.1053625647 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 13 |
Hb_001014_210 |
0.1054292364 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 14 |
Hb_005463_110 |
0.105665941 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 15 |
Hb_001662_100 |
0.1103569398 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 16 |
Hb_000140_290 |
0.1103838417 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 17 |
Hb_000890_060 |
0.1107362837 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 18 |
Hb_000252_050 |
0.1111370442 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 19 |
Hb_000322_080 |
0.1113521766 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 20 |
Hb_000085_450 |
0.1114576362 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |