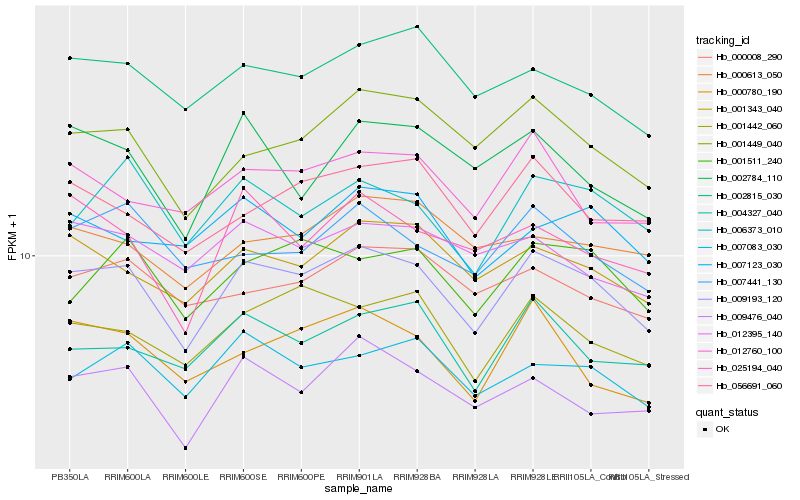
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009193_120 |
0.0 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 2 |
Hb_001343_040 |
0.0783480726 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 3 |
Hb_001449_040 |
0.0808378541 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001442_060 |
0.0833739967 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006373_010 |
0.0839181785 |
- |
- |
Potassium transporter 11 [Gossypium arboreum] |
| 6 |
Hb_007083_030 |
0.0844992785 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 7 |
Hb_001511_240 |
0.0852940281 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 8 |
Hb_009476_040 |
0.0870664849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007441_130 |
0.0873439247 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_002815_030 |
0.0876492906 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 11 |
Hb_004327_040 |
0.0878736287 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 12 |
Hb_056691_060 |
0.0905667235 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012760_100 |
0.0918355365 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 14 |
Hb_007123_030 |
0.093203558 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000008_290 |
0.0933469849 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 16 |
Hb_000780_190 |
0.0937956773 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 17 |
Hb_025194_040 |
0.0938174104 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 18 |
Hb_012395_140 |
0.0940131483 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 19 |
Hb_002784_110 |
0.0944855853 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 20 |
Hb_000613_050 |
0.0954030885 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |