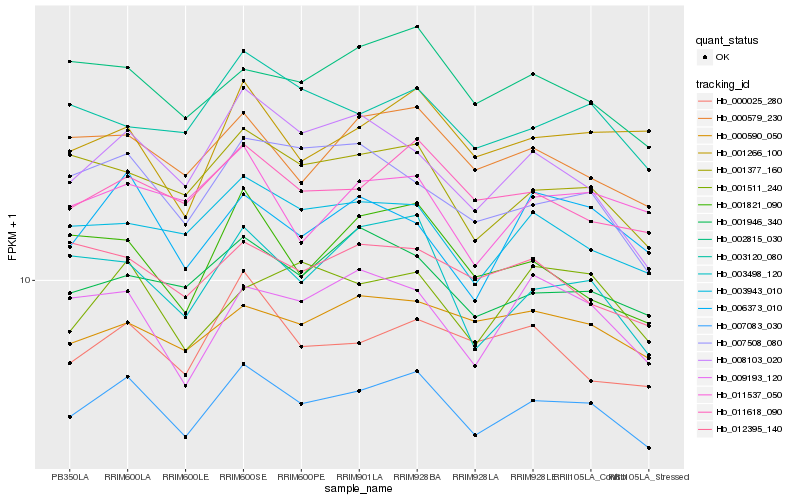
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007083_030 |
0.0 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 2 |
Hb_011618_090 |
0.0677120773 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_001377_160 |
0.0708337355 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 4 |
Hb_000579_230 |
0.0718293453 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 5 |
Hb_003943_010 |
0.0724339475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002815_030 |
0.0781679442 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 7 |
Hb_001946_340 |
0.0797300033 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000590_050 |
0.08087654 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 9 |
Hb_003498_120 |
0.0818718201 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 10 |
Hb_003120_080 |
0.0822868475 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007508_080 |
0.0834790143 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 12 |
Hb_000025_280 |
0.0842543891 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 13 |
Hb_009193_120 |
0.0844992785 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 14 |
Hb_006373_010 |
0.0849524224 |
- |
- |
Potassium transporter 11 [Gossypium arboreum] |
| 15 |
Hb_008103_020 |
0.0856428362 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 16 |
Hb_001511_240 |
0.0856950201 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 17 |
Hb_001266_100 |
0.0859154837 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 18 |
Hb_001821_090 |
0.0863183873 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 19 |
Hb_012395_140 |
0.0867525447 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 20 |
Hb_011537_050 |
0.0869483465 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |