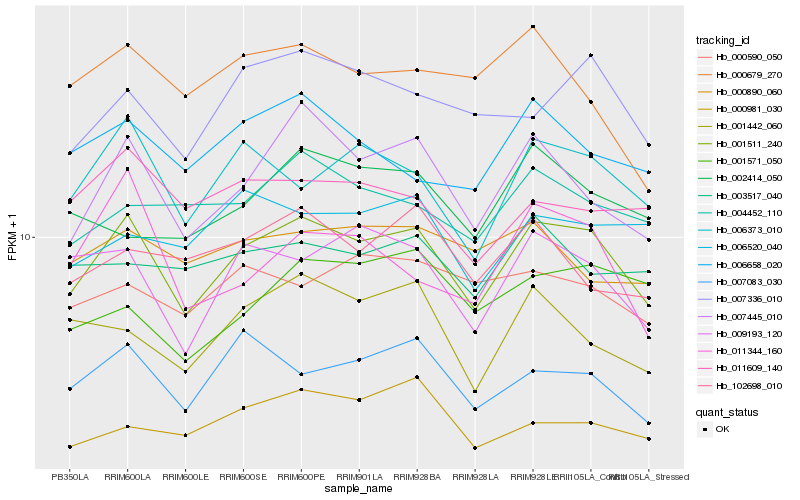
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 2 |
Hb_007445_010 |
0.0738798869 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_006373_010 |
0.0813294773 |
- |
- |
Potassium transporter 11 [Gossypium arboreum] |
| 4 |
Hb_009193_120 |
0.0852940281 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 5 |
Hb_007083_030 |
0.0856950201 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 6 |
Hb_007336_010 |
0.0879506665 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 7 |
Hb_004452_110 |
0.0933015492 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001442_060 |
0.0934743713 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006658_020 |
0.0950021569 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 10 |
Hb_000981_030 |
0.0965985821 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 11 |
Hb_000590_050 |
0.0994736728 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 12 |
Hb_102698_010 |
0.0996168801 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_011609_140 |
0.0997301243 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 14 |
Hb_006520_040 |
0.1001133319 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000890_060 |
0.1006302457 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 16 |
Hb_003517_040 |
0.1008083696 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001571_050 |
0.1022155692 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 18 |
Hb_011344_160 |
0.1023541464 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 19 |
Hb_002414_050 |
0.1027916832 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 20 |
Hb_000679_270 |
0.1036685097 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |