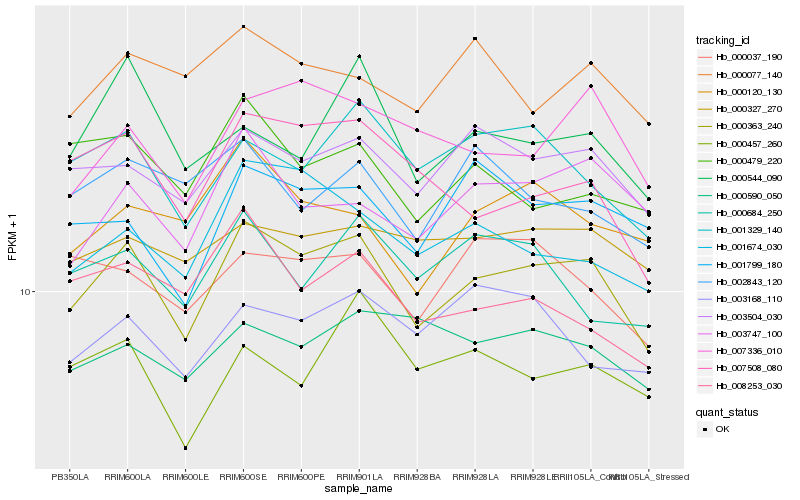
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_240 |
0.0 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |
| 2 |
Hb_003504_030 |
0.0744406101 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001329_140 |
0.0831564881 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 4 |
Hb_001674_030 |
0.0841358802 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 5 |
Hb_007336_010 |
0.0846967789 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 6 |
Hb_003747_100 |
0.0856475967 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_007508_080 |
0.0861829265 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 8 |
Hb_001799_180 |
0.0891229403 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 9 |
Hb_000037_190 |
0.0893791573 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 10 |
Hb_000120_130 |
0.0894090795 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 11 |
Hb_000327_270 |
0.089983909 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_008253_030 |
0.0922381563 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 13 |
Hb_000544_090 |
0.0925739677 |
- |
- |
PREDICTED: golgin candidate 2 [Jatropha curcas] |
| 14 |
Hb_000479_220 |
0.0930258372 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 15 |
Hb_000077_140 |
0.0931272267 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 16 |
Hb_003168_110 |
0.0938153664 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 17 |
Hb_000684_250 |
0.0946645024 |
- |
- |
PREDICTED: uncharacterized protein LOC105642172 [Jatropha curcas] |
| 18 |
Hb_000590_050 |
0.0947271089 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 19 |
Hb_002843_120 |
0.0965759214 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000457_260 |
0.0972628285 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |