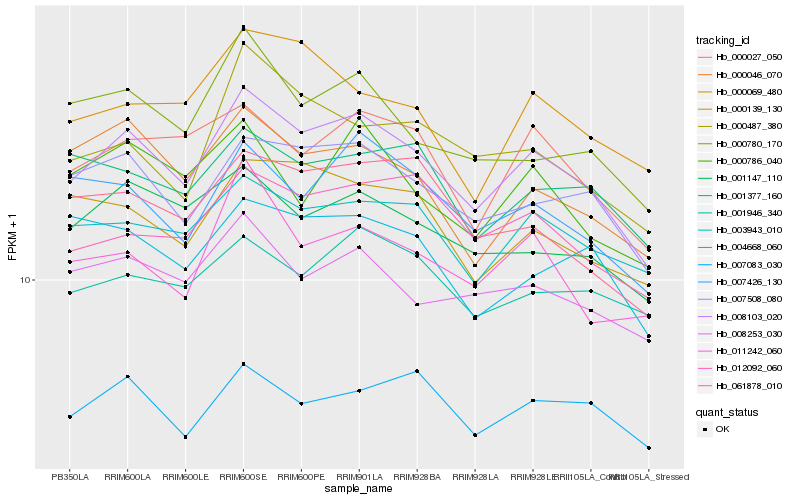
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008103_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 2 |
Hb_007508_080 |
0.0659782795 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 3 |
Hb_000027_050 |
0.0770811416 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003943_010 |
0.0776939718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007426_130 |
0.0783081127 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 6 |
Hb_001147_110 |
0.0785231417 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 7 |
Hb_008253_030 |
0.0801451422 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 8 |
Hb_000046_070 |
0.08123466 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 9 |
Hb_000780_170 |
0.0813157904 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 10 |
Hb_000139_130 |
0.0828791152 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 11 |
Hb_061878_010 |
0.0832312253 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 12 |
Hb_012092_060 |
0.0836069809 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 13 |
Hb_007083_030 |
0.0856428362 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000786_040 |
0.0858966924 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 15 |
Hb_011242_060 |
0.0862764329 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 16 |
Hb_000487_380 |
0.0876402657 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001946_340 |
0.0889320642 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001377_160 |
0.0898892187 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 19 |
Hb_004668_060 |
0.0900605394 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000069_480 |
0.0905469078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |