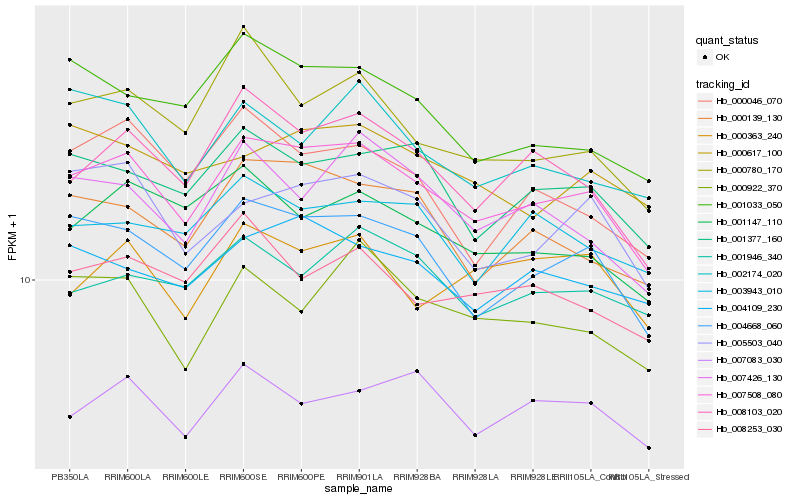
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007508_080 |
0.0 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 2 |
Hb_004668_060 |
0.0533350754 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008103_020 |
0.0659782795 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 4 |
Hb_000139_130 |
0.0691613079 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 5 |
Hb_001377_160 |
0.0724218759 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 6 |
Hb_004109_230 |
0.0765542858 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_005503_040 |
0.0778155405 |
- |
- |
PREDICTED: AMP deaminase [Jatropha curcas] |
| 8 |
Hb_007426_130 |
0.0783342482 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 9 |
Hb_001147_110 |
0.0797317002 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 10 |
Hb_000922_370 |
0.0818231578 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 11 |
Hb_008253_030 |
0.0822029755 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 12 |
Hb_003943_010 |
0.0828326848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001033_050 |
0.0831845259 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 14 |
Hb_000046_070 |
0.0833045039 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 15 |
Hb_001946_340 |
0.0833664838 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_007083_030 |
0.0834790143 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 17 |
Hb_000780_170 |
0.0840538453 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 18 |
Hb_000363_240 |
0.0861829265 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |
| 19 |
Hb_002174_020 |
0.086442755 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_000617_100 |
0.087044618 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |