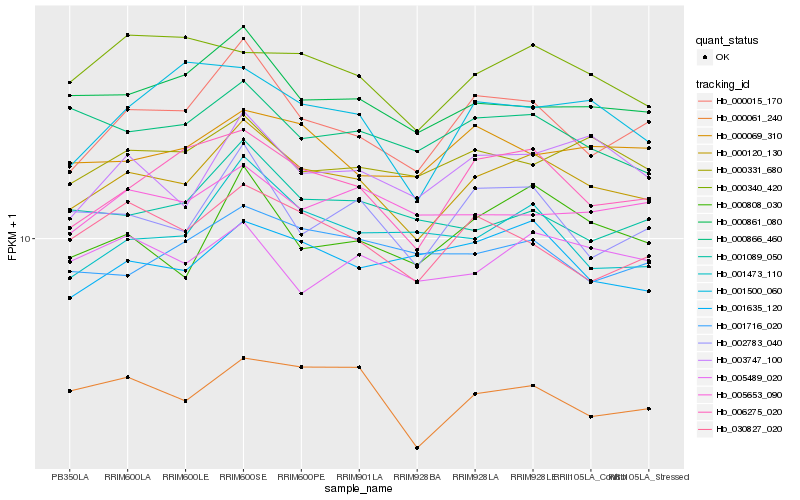
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_130 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 2 |
Hb_006275_020 |
0.0618905826 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000015_170 |
0.0636219887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000861_080 |
0.0708005363 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 5 |
Hb_000808_030 |
0.0740603613 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 6 |
Hb_003747_100 |
0.0755341692 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002783_040 |
0.0762708312 |
transcription factor |
TF Family: IWS1 |
- |
| 8 |
Hb_000331_680 |
0.0782139014 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 9 |
Hb_030827_020 |
0.0802097288 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 10 |
Hb_000069_310 |
0.0811275489 |
- |
- |
ubiquitin ligase [Cucumis melo subsp. melo] |
| 11 |
Hb_001473_110 |
0.0819077847 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 12 |
Hb_001089_050 |
0.0824051616 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_000866_460 |
0.0824074383 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_005653_090 |
0.0829693812 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 15 |
Hb_001635_120 |
0.0831295746 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 16 |
Hb_000340_420 |
0.0835242358 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_000061_240 |
0.0838010888 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_001716_020 |
0.0842310494 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 19 |
Hb_005489_020 |
0.0855694085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001500_060 |
0.0856255487 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |