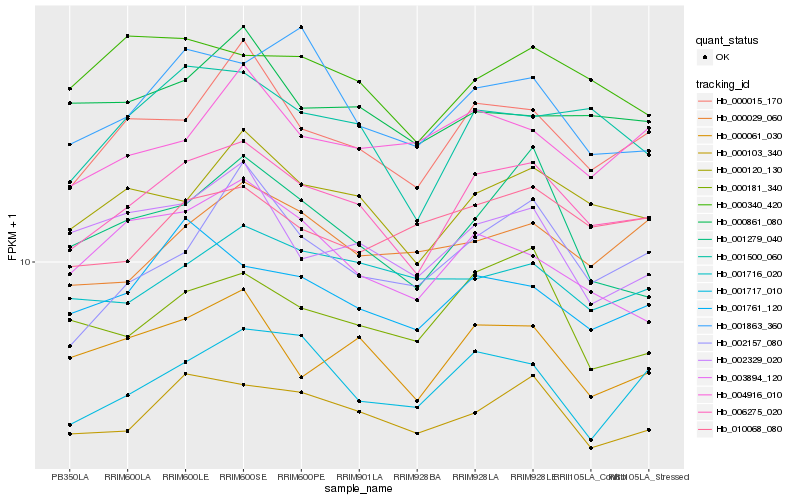
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006275_020 |
0.0 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 2 |
Hb_000120_130 |
0.0618905826 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 3 |
Hb_001500_060 |
0.0646729345 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |
| 4 |
Hb_000015_170 |
0.0658788912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000103_340 |
0.069038926 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000061_030 |
0.0755762627 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |
| 7 |
Hb_001761_120 |
0.0832421403 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 8 |
Hb_001717_010 |
0.0874593849 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 9 |
Hb_000340_420 |
0.087992317 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_000181_340 |
0.0884549672 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 11 |
Hb_001279_040 |
0.0896798212 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 12 |
Hb_000029_060 |
0.0900432507 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 13 |
Hb_002157_080 |
0.0902307783 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 14 |
Hb_010068_080 |
0.0909183719 |
- |
- |
PREDICTED: RNA-binding protein rsd1-like [Jatropha curcas] |
| 15 |
Hb_001716_020 |
0.0912210408 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 16 |
Hb_001863_360 |
0.0919950904 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 17 |
Hb_004916_010 |
0.0921178537 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 18 |
Hb_002329_020 |
0.0921575143 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 19 |
Hb_000861_080 |
0.0924570552 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 20 |
Hb_003894_120 |
0.092626554 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |