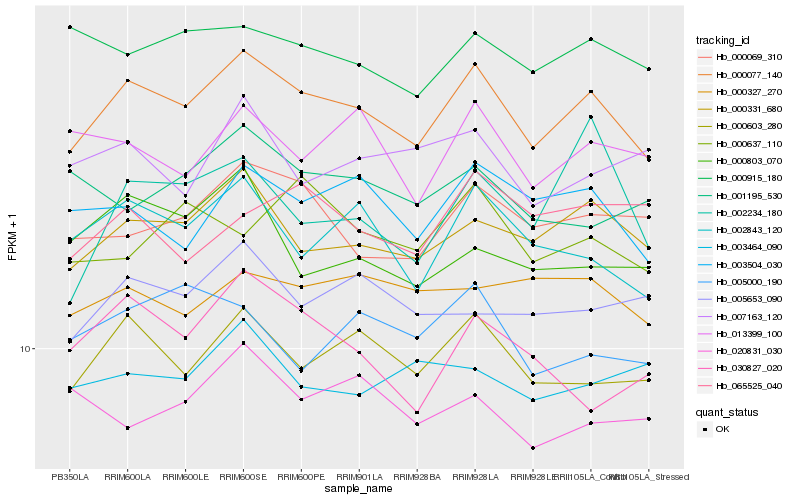
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_140 |
0.0 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 2 |
Hb_000331_680 |
0.0547019356 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_000603_280 |
0.0567364005 |
- |
- |
PREDICTED: uncharacterized protein LOC105638082 [Jatropha curcas] |
| 4 |
Hb_002843_120 |
0.063217069 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003504_030 |
0.0660167796 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000069_310 |
0.0679501629 |
- |
- |
ubiquitin ligase [Cucumis melo subsp. melo] |
| 7 |
Hb_000915_180 |
0.0738334417 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |
| 8 |
Hb_005653_090 |
0.0743372971 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 9 |
Hb_013399_100 |
0.0748499616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000327_270 |
0.0753944179 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_007163_120 |
0.0805176208 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 12 |
Hb_000803_070 |
0.0808816621 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 13 |
Hb_005000_190 |
0.0811814515 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |
| 14 |
Hb_001195_530 |
0.0815915909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000637_110 |
0.0816471561 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 16 |
Hb_003464_090 |
0.0818412005 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 17 |
Hb_065525_040 |
0.0824920801 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 18 |
Hb_002234_180 |
0.0828176809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_030827_020 |
0.0830122452 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 20 |
Hb_020831_030 |
0.0833335895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |