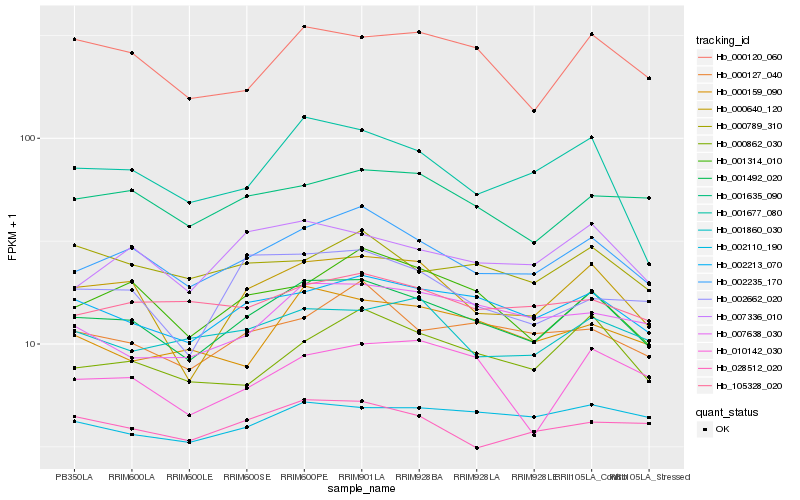
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001492_020 |
0.0 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 2 |
Hb_002235_170 |
0.0491990463 |
- |
- |
hypothetical protein CISIN_1g010208mg [Citrus sinensis] |
| 3 |
Hb_002213_070 |
0.0575567032 |
- |
- |
epsilon-adaptin family protein [Populus trichocarpa] |
| 4 |
Hb_000640_120 |
0.0622874516 |
- |
- |
PREDICTED: uncharacterized protein LOC105642625 [Jatropha curcas] |
| 5 |
Hb_007336_010 |
0.0739916814 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 6 |
Hb_000120_060 |
0.0741854846 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 7 |
Hb_001314_010 |
0.0764635489 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 1-like [Jatropha curcas] |
| 8 |
Hb_000862_030 |
0.0766472622 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000127_040 |
0.0780635541 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 10 |
Hb_001635_090 |
0.0802360786 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 11 |
Hb_028512_020 |
0.0825959439 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001860_030 |
0.0847573281 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 13 |
Hb_001677_080 |
0.0855765413 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000789_310 |
0.0856278006 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT1 [Jatropha curcas] |
| 15 |
Hb_010142_030 |
0.0874683726 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_002662_020 |
0.0876083913 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 17 |
Hb_007638_030 |
0.0877535491 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 18 |
Hb_002110_190 |
0.0879451495 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27460 [Jatropha curcas] |
| 19 |
Hb_000159_090 |
0.0891247305 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 20 |
Hb_105328_020 |
0.0893024094 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |