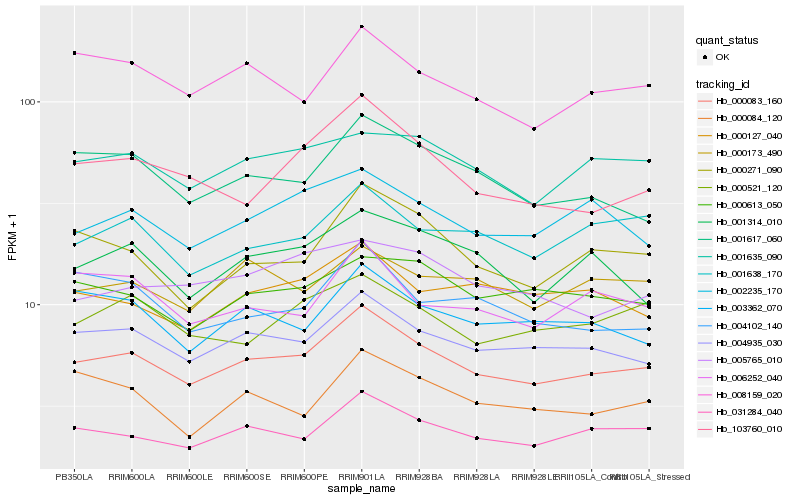
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_160 |
0.0 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 2 |
Hb_004935_030 |
0.0521477992 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 3 |
Hb_031284_040 |
0.0555538722 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 4 |
Hb_000271_090 |
0.0749354696 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_001617_060 |
0.0752544782 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 6 |
Hb_001635_090 |
0.0764344514 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 7 |
Hb_002235_170 |
0.0766307103 |
- |
- |
hypothetical protein CISIN_1g010208mg [Citrus sinensis] |
| 8 |
Hb_008159_020 |
0.0768681421 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 9 |
Hb_003362_070 |
0.0781757017 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 10 |
Hb_001638_170 |
0.0786536054 |
- |
- |
hypothetical protein Csa_3G118170 [Cucumis sativus] |
| 11 |
Hb_103760_010 |
0.0790484198 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 12 |
Hb_000613_050 |
0.0803076317 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000521_120 |
0.0806509296 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000127_040 |
0.0813569161 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 15 |
Hb_001314_010 |
0.0818585073 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 1-like [Jatropha curcas] |
| 16 |
Hb_000173_490 |
0.0822928007 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006252_040 |
0.0838713967 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_005765_010 |
0.084804036 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000084_120 |
0.0851220674 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g36240 [Jatropha curcas] |
| 20 |
Hb_004102_140 |
0.0857676399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |